Figure 8. 15N relaxation parameters for consensus-HD and engrailed-HD.
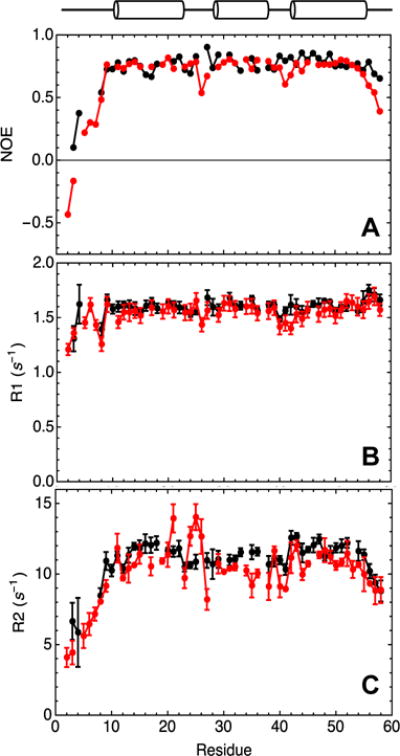
A) Heteronuclear NOE, B) R1, and C) R2 parameters for consensus-HD (black) and engrailed-HD (red) shown as a function of sequence position. Errors shown for R1 and R2 parameters for each residue are standard errors of fits. Overlapping peaks (L16, R18, E22, Q33, E37, K46 for engrailed-HD; T7, L19, E30, L34, K37, R53 for consensus-HD) and peaks with low signal intensity (R5 and T6 for consensus-HD) were excluded from analysis. Helix positions are based on engrailed-HD crystal structure. Sequence numbering as in Figure 3. Conditions: 25 mM sodium phosphate, 150 mM NaCl, pH 7.0, 20°C.
