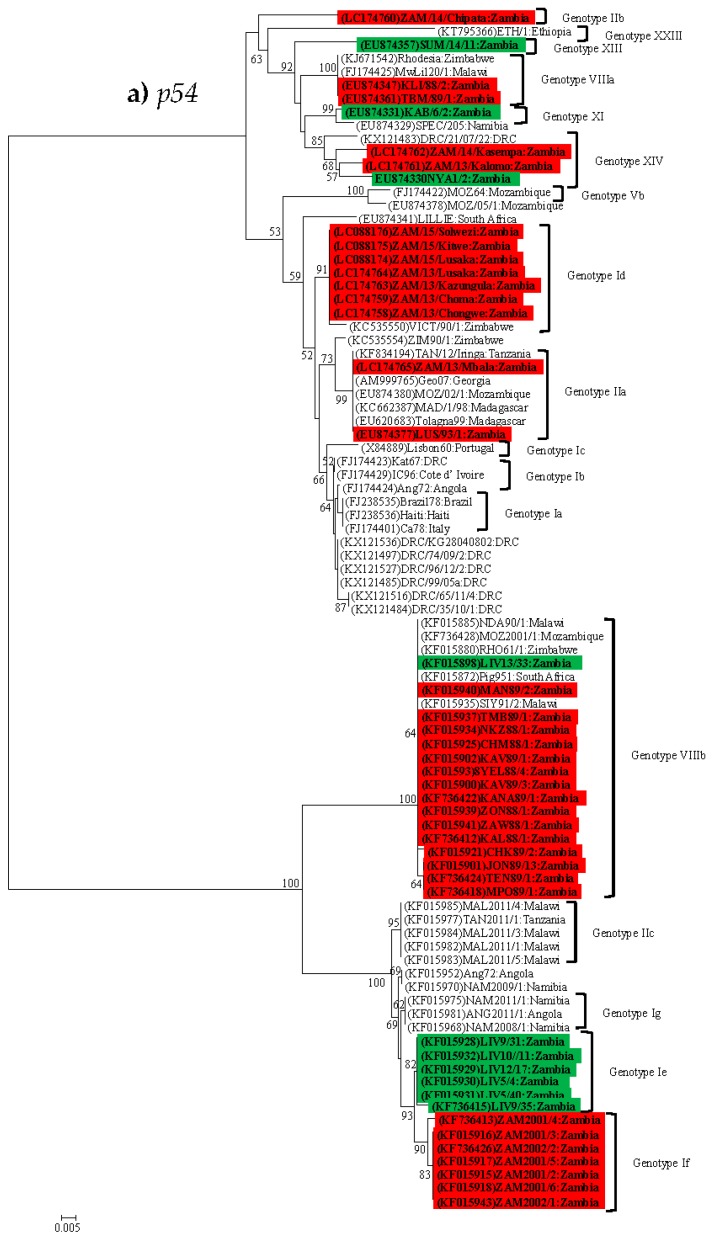
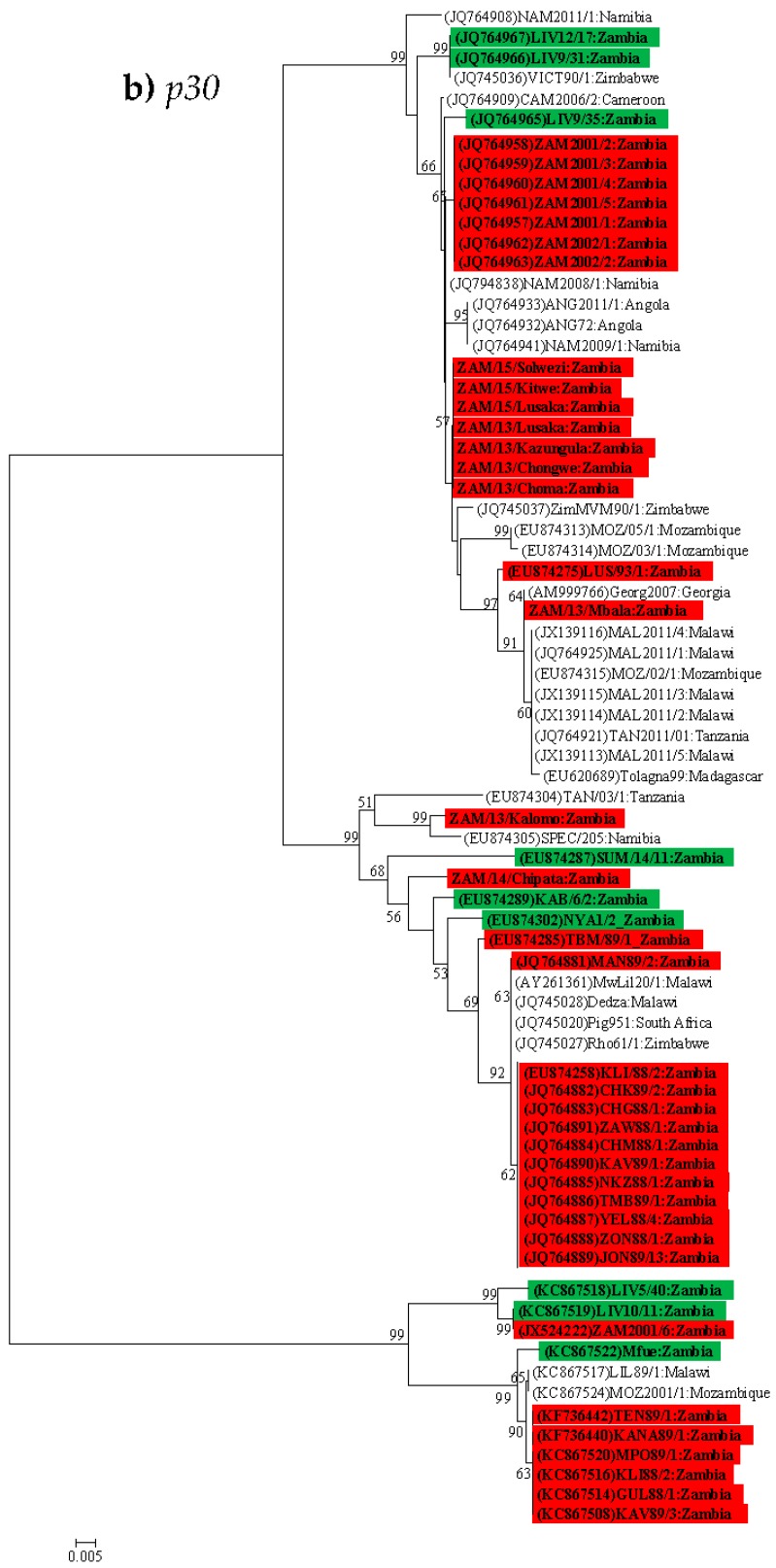
Figure 4.
Phylogenetic relationships of the p54 (a) and p30 (b) genes of ASFVs detected in pigs and soft ticks in Zambia. Analysis was based on 483 bp of the p54 and 528 bp of the p30 genes. The evolutionary history was inferred using the Minimum Evolution method with evolutionary distances being computed using the p-distance method. The ME tree was searched using the close-neighbor-interchange (CNI) algorithm at a search level of 1. The neighbor-joining algorithm was used to generate the initial tree. Numbers at branch nodes indicate bootstrap values ≥50%. The GenBank accession no. of strains included in the analysis are indicated in parenthesis. Virus strains characterized in the present study detected in domestic pigs are shaded in red while those from ticks are shaded in green. Bar, number of substitutions per site.