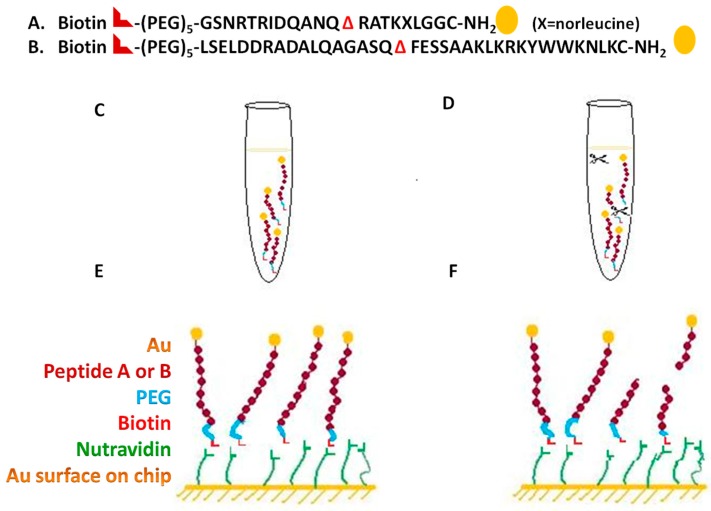
Figure 3.
Schematics of the endopeptidase assay design for SPR. The change in the reflective index directly corresponds to the change in mass of the peptide substrate at the chip surface. (A) Indicated substrate sequence for the botulinum neurotoxin A, the peptide A incorporated with the cleavage site Q–R, (Δ represents the cleavage site); (B) The indicated substrate sequence for the botulinum neurotoxin B, the substrate incorporated the cleavage site Q–F (Δ represents the cleavage site); (C) Control (uncleaved peptide A or B) for the SPR sample included 3pM peptide A or B; (D) 3 pM peptide A or B sample spiked with different concentrations of botulinum neurotoxin A or B light chain in the concentration range 6.6 µg/mL to 6.6 pg/mL; (E) Corresponded to the control (uncleaved peptide A or B) at the SPR Au chip surface. Biotin binds with the nutravidin at the Au chip surface; (F) Corresponds to the sample dose-dependent cleaved peptide samples bound through biotin at the Au chip surface to nutravidin.