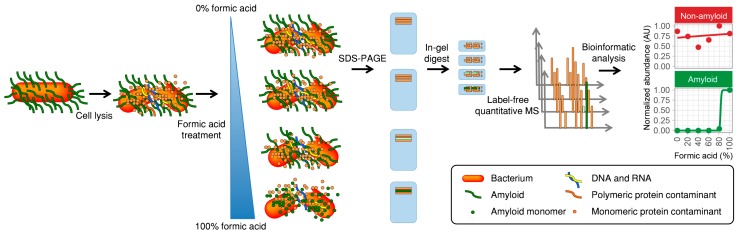
Figure 1.
Direct identification of functional amyloid proteins using label-free quantitative (LFQ) Liquid chromatography–tandem mass spectrometry (LC-MS/MS). The sample is lysed and divided into aliquots that are lyophilized and treated with either 0%, 20%, 40%, 60%, 80%, or 100% formic acid. The samples are then lyophilized, dissolved in reducing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, and subjected to short run SDS-PAGE. The amyloid proteins can only enter the gel if they have been pretreated with concentrated formic acid and are therefore only present in these samples. In-gel digestion is carried out with trypsin, and samples analyzed by label-free quantitative LC-MS/MS using MaxQuant and the MaxLFQ algorithm. The data is finally analyzed for each protein using an automated script, and positive amyloid candidates are identified based on their abundance profiles with respect to the formic acid concentration.