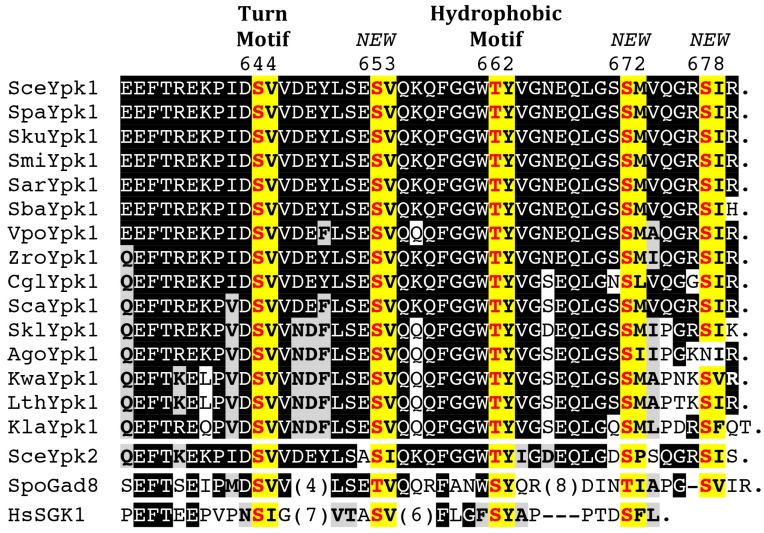
Figure 5.
Conservation of sites phosphorylated by TORC2 in Ypk1. The C-terminal end of Saccharomyces cerevisiae (Sce) Ypk1 (top line) was aligned with the corresponding segment of Ypk1 orthologs from sensu stricto Saccharomyces species S. paradoxus (Spa), S. kudriavzevii (Sku), S. mikatae (Smi), S. arboricola (Sar), and S. bayanus (Sba), more divergent species Vanderwaltozyma polyspora (Vpo), Zygosaccharomyces rouxii (Zro), Candida glabrata (Cgl), S. castellii (Sca), S. kluyveri (Skl), Ashbya gossypii (Ago), Kluveromyces waltii (Kwa), Lachancea thermotolerans (Lth), and Kluveromyces lactis (Kla), SceYpk2, Schizosaccharomyces pombe (Spo), and Homo sapiens (Hs) SGK1 (bottom line). Identities (white letters on black boxes) and standard conservative substitutions (bold letters on grey boxes) shared between SceYpk1 and another ortholog are indicated. One-residue gaps (hyphens) and insertions of the indicated length (parentheses) were introduced to maximize alignment of the most distant orthologs. Period (.) indicates the end of the open-reading-frame. The proximal seven-residue insert in HsSGK1 contains two additional potential mTORC2 sites (KSPDSVL). Sequence sources can be found in [39].