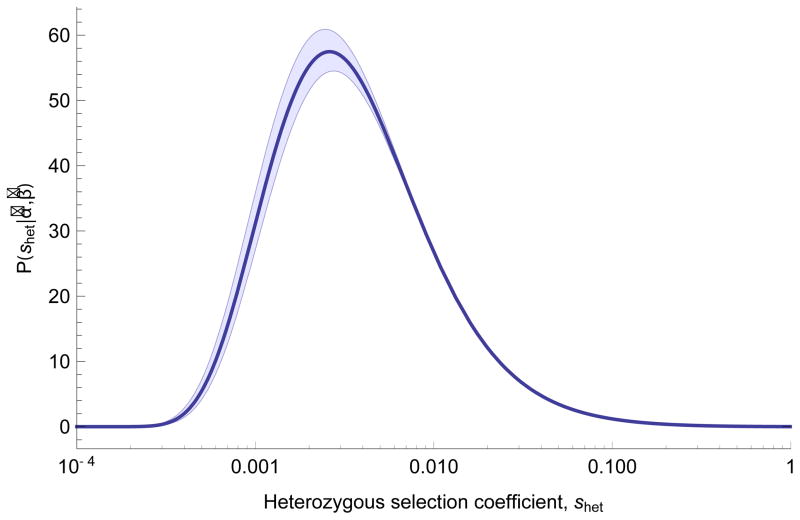
Figure 1.
Inferred distribution of fitness effects for heterozygous loss of gene function. Estimates of parameters (α̂, β̂) from maximum likelihood fit to the observed distribution of PTV counts across 15,998 genes in terciles of mutation rate, assuming shet ~ IG(α, β). Shaded areas show 95% CI obtained from 100 bootstrapping replicates, intended to quantify the influence of sampling noise in the data set on parameter inference, with fixed estimates of local mutation rate.