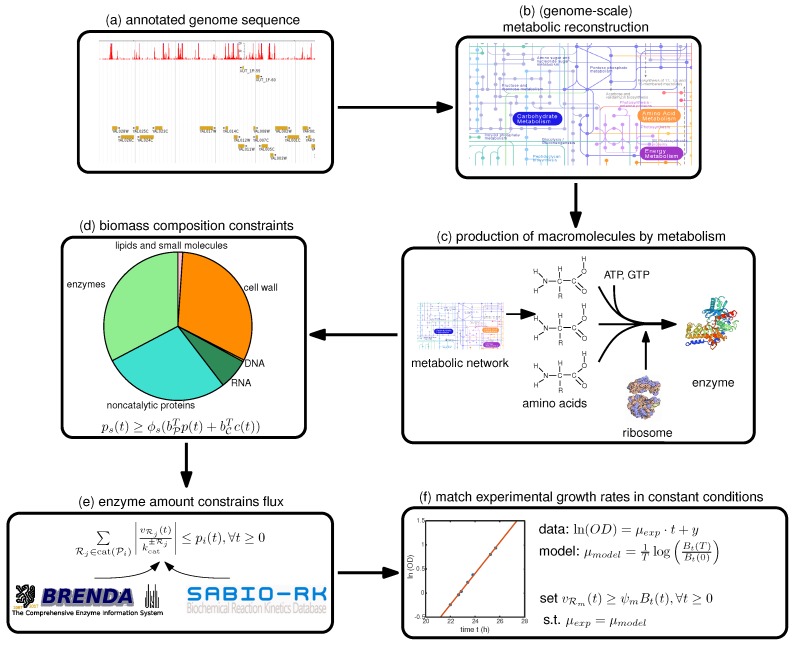
Figure 1.
Protocol for generating a deFBA model. From an annotated genome sequence (a) of the organism of interest the metabolic network (b) is reconstructed following instructions in [15]. Given the gene-reaction mapping and the annotated genome sequence, the enzymes and ribosomes (c); and their synthesis reactions are added to the stoichiometric matrix (see Section 4). Next, the biomass composition constraints (d) should be set up using information from the biomass objective function of the metabolic network model (see Section 5). Then reaction turnover rates (e) sourced from literature and online databases should be added (see Section 6). Lastly but most importantly, the deFBA model should be fine tuned to match experimental growth rates (f) obtained in the laboratory (see Section 7). Images retrieved from: (a) http://goo.gl/aBNfPz [16,17]; (b) http://www.genome.jp/kegg-bin/show_pathway?map01100 [18]; (c) http://www.genome.jp/kegg-bin/show_pathway?map01100 [18], http://pdb101.rcsb.org/motm/10 [19], https://swissmodel.expasy.org/repository/uniprot/P04806 [20].