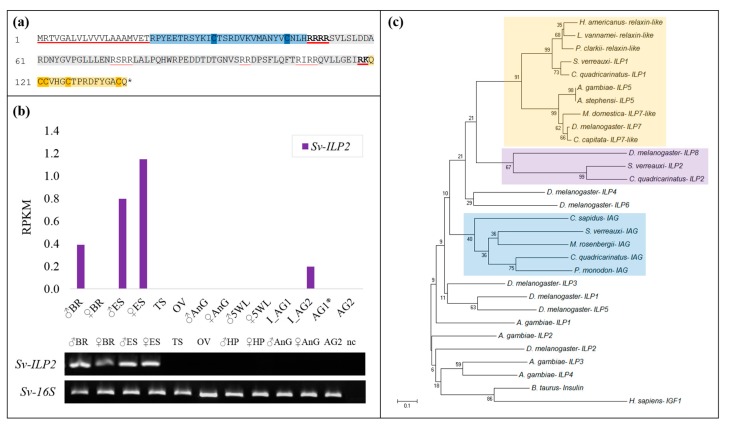
Figure 1.
Spatial expression and phylogeny of Sv-ILP2: (a) sequence of Sv-ILP2, the signal peptide is underlined in red and the B-(blue) and A-(orange) chains boxed with the cysteine core of each highlighted. The C-peptide is shown in grey with the predicted Arg-C cleavage sites shown, with those predicted to generate the mature hormone underlined in red. (b) Transcriptomic spatial expression of Sv-ILP2 quantified as reads per kilobase per million reads (RPKM) for male and female brain (BR), eyestalk (ES), gonads (TS and OV), antennal gland (AnG), and fifth walking leg (5WL), immature androgenic glands (I_AG1 and I_AG2), and mature androgenic glands (AG1* and AG2, where * indicates a hypertrophied gland). Validated with spatial RT-PCR analyses, with the removal of the immature AGs, 5WL, and AG1*, and the addition of male and female hepatopancreas (HP). Negative control (NC) in the fifteenth lane, 16S as positive control. (c) Neighbour-joining phylogram of Sv- and Cq-ILP1 and ILP2 with a range of ILPs from model species: Anopheles ILP1-5, Drosophila Dilp1-8, decapod insulin-like androgenic gland peptides (IAGs), and bovine insulin and human IGFI. Bootstrap values are shown at each node and were performed with 1000 replicates. Scale bar indicates number of amino acid substitutions per site. IAG cluster boxed in blue, Dilp7/relaxin-like cluster in yellow, and novel decapod ILP2s in purple.