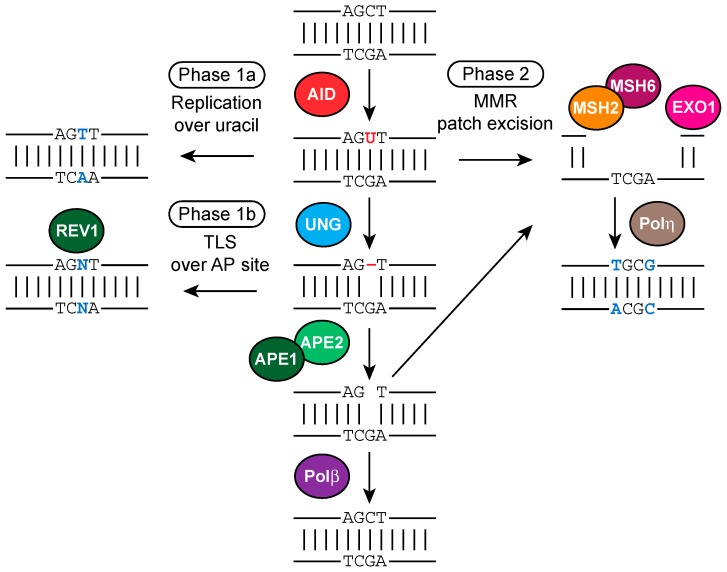
Figure 7.
Molecular mechanism of Phases 1 and 2 SHM, involving MMR and BER proteins. Processing of AID-instigated uracils (shown in red font color) determine the somatic hypermutation (SHM) spectrum (mutations are shown in blue font color). Replication over uracils result in C-to-T/G-to-A transition mutations (Phase 1a SHM). Uracil removal by uracil-DNA-glycosylase (UNG) followed by replication over the apurinic/apyrimidinic (AP) site (shown as red dash) by translesion synthesis (TLS) DNA polymerases such as REV1 result in transition or transversion mutations are C/G base pairs (bp) (Phase 1b SHM). Recognition of AID-instigated U:G mismatches by the mismatch repair (MMR) pathway components Mut S homolog 2 (MSH2) and MSH6 and recruitment of DNA exonuclease 1 (EXO1) lead to patch excision, and subsequent recruitment of DNA polymerase η (Polη), which is error-prone particularly at A and T bps, results in A/T mutations (Phase 2 SHM). AP sites can be cleaved by AP endonuclease 1/2 (APE1/2), the resulting nicks may serve as entry points for EXO1 and thereby stimulate Phase 2 mutations. Alternatively, DNA Polβ triggers faithful repair of AID-induced lesions.