Figure 2.
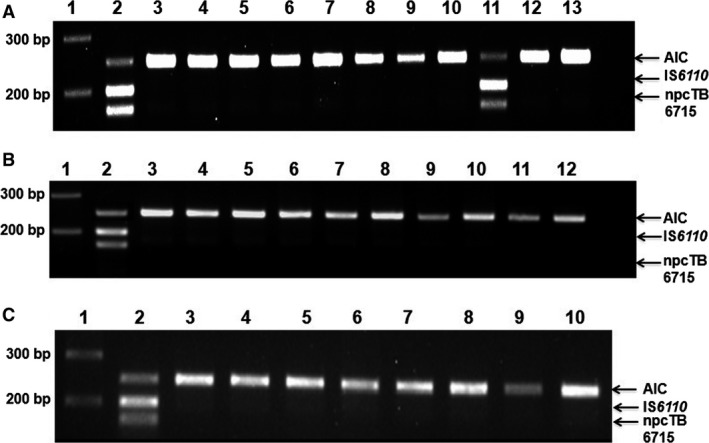
Specificity of the mPCR assay analysed with 4% agarose gel‐electrophoresis. The template DNA was 10 ng of MTB H37Rv genomic DNA per reaction. Primers for IS 6110 and npcTB_6715 were used at 0.1 μM and 0.5 μM, respectively. (A) MTB complex and NTM, Lane 1: 100 bp DNA ladder, Lane 2: MTB H37Rv, Lane 3: M. avium, Lane 4: M. abscessus, Lane 5: M. fortuitum, Lane 6: M. fortuitum, Lane 7: M. gordonae, Lane 8: M. scroferaceum, Lane 9: M. intracellulare. Lane 10: M. kansasii, Lane 11: MTB H37Ra, Lane 12: M. malmoense, Lane 13: Negative control (B) NTM and lower respiratory pathogens, Lane 1: 100 bp DNA ladder, Lane 2: MTB H37Rv, Lane 3: M. marinum, Lane 4: M. chelonae, Lane 5: Staphylococcus aureus, Lane 6: Streptococcus pneumoniae, Lane 7: Klebsiella pneumoniae, Lane 8: Pseudomonas aeruginosa, Lane 9: Haemophilus influenza. Lane 10: Haemophilus parainfluenza, Lane 11: Moraxella catarrhalis, Lane 12: Negative control (C) normal flora bacterial strains. Lane 1: 100 bp DNA ladder, Lane 2: MTB H37Rv, Lane 3: Escherichia coli, Lane 4: Bifidobacterium bifidum, Lane 5: Corynebacteria sp., Lane 6: Lactobacillus sp, Lane 7: Neisseria meningitidis, Lane 8: Staphylococcus epidermidis, Lane 9: Coagulase Negative Streptococcus, Lane 10: Negative control.
