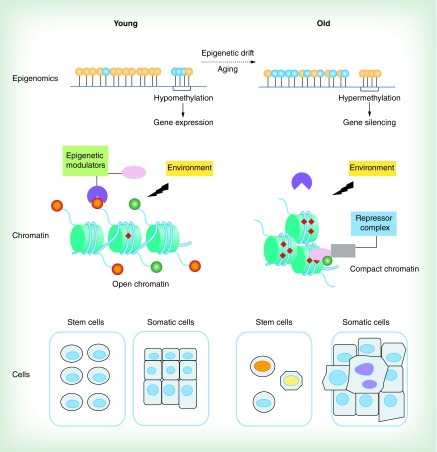
Figure 1. . A model of epigenetic drift-induced cellular phenotypic plasticity attenuation during aging.
Aging is associated with gradual deteriorations of epigenomic markers (known as epigenetic drift) such as altered methylomic patterns (top panel). Aging results in global hypomethylation in the DNA genome but induces hypermethylation in the regulatory regions of certain key genes (orange circle represents methylated CpG island and blue circle represents unmethylated CpG island, top panel). As depicted in the middle panel, ‘normal’ chromatin states are found throughout the genome in young cells. A hypothetical gene expression is activated and well controlled by the regulatory epigenetic machinery including regulations of epigenetic modulators and active epigenetic landmarks (dark orange circle, methylated histone tail; green circle, acetylated histone tail; red diamond, methylated CpG island) in response to environmental factors (middle left). With age proceeding, the regulatory epigenetic machinery becomes gradually deregulated. The aged epigenome fails to respond to environmental factors leading to an altered epigenetic state with a loss of active epigenetic markers and DNA hypermethylation, resulting in compacted chromatin, repressor complex recruitment and subsequent gene silencing (middle right). At the cellular level, young stem cells have relatively identical epigenomes (bottom left) and show robust self-renewal function. During aging, accumulated epigenetic deteriorations lead to epigenomic mosaicism in stem cells resulting in reduced regenerative capacity of stem cells and exhaustion of the stem cell pool (bottom right). In somatic cells, massive epigenetic lesions as well as accumulative genetic mutation with age lead to emergence of hyperproliferative somatic clones (represented as enlarged and irregular cells that have invaded normal tissue boundary, bottom right) in affected tissues that contributes to tumorigenesis and other age-associated diseases.