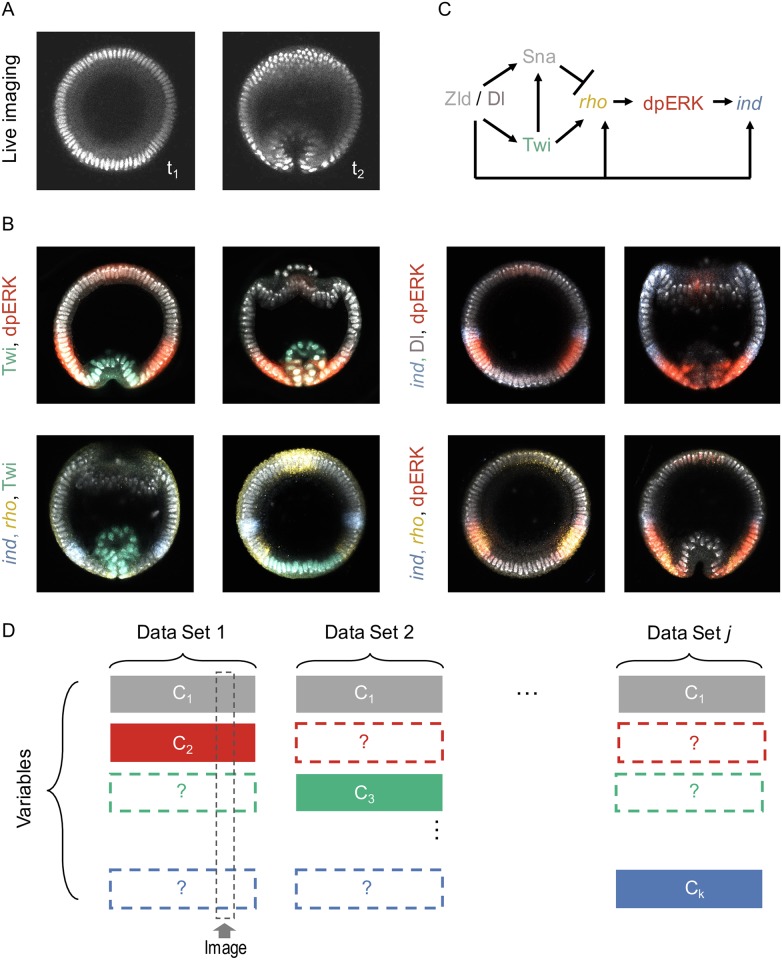
Fig 1. Stating the problem of data fusion.
(A-B) Example datasets of molecular signals and morphology during the DV patterning of Drosophila embryo; all images are collected from optical cross-section along the DV axis, ∼ 15% from the posterior pole of the embryo. (A) Frames from a live imaging movie, showing positions of nuclei during the early stages of gastrulation. (B) Images of fixed embryos, stained with probes and antibodies revealing the spatial patterns of nuclear Dl (pink), Twi (green), dually phosphorylated ERK (red), and transcripts of rho (yellow) and ind (blue). (C) A fragment of a DV patterning network in the early Drosophila embryo. (D) Data fusion as a matrix completion problem: Each row corresponds to a variable, e.g. nuclear positions, gene expression levels, time stamp, revealed by visualizing different molecular or cellular components, nuclei, transcripts, or protein phosphorylation. Each column of the matrix corresponds to an image giving access to some of the states through various channels. The remaining states, labeled with a question mark, must be estimated from other datasets.