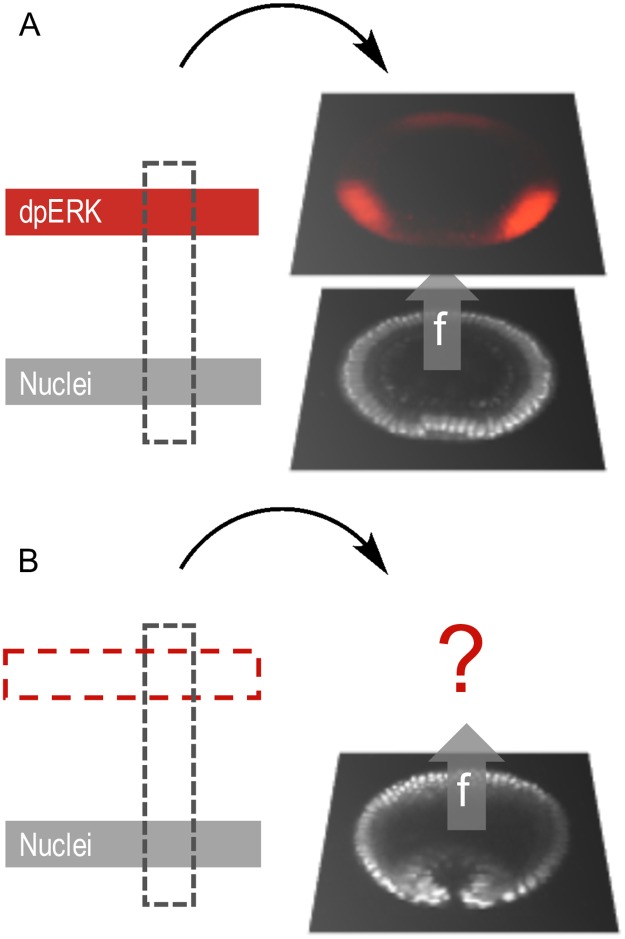
Fig 2. Learning a mapping from a common channel.
An experimental image can be decomposed into various channels. E.g., red: dpERK, visualized with a phosphospecific antibody, gray: nuclei, visualized through either DAPI (in fixed images) or Histone-RFP (in live imaging). The training ensemble of labeled images (A) is used to predict the labels on a set of unlabeled images (B) using common information, the morphology obtained through the nuclei signal in this case. Morphological proximity yields similar labels.