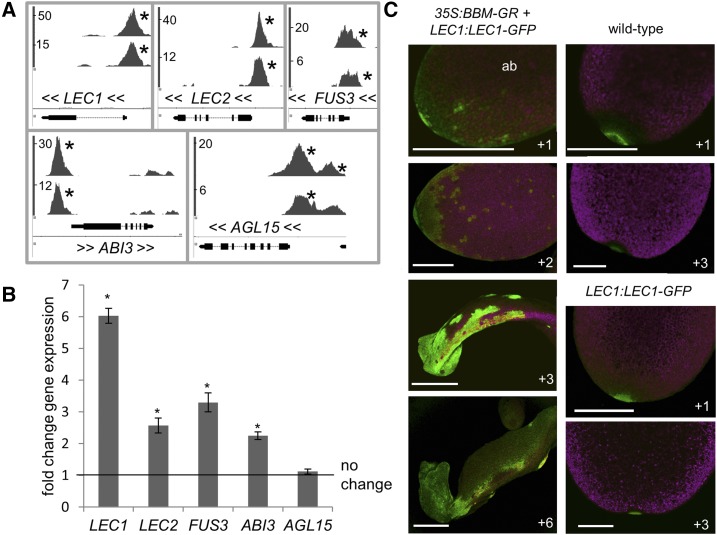
Figure 1.
BBM binds and transcriptionally regulates LAFL genes. A, ChIP-seq BBM-binding profiles for embryo-expressed genes: 35S:BBM-GFP (top profiles) and BBM:BBM-YFP (bottom profiles). x axis, Nucleotide positions (TAIR 10 annotation; black bars indicate exons, and lines indicate introns); y axis, ChIP-seq score (fold enrichment of the BBM-GFP/YFP ChIP to the control ChIP), as calculated by the CSAR package in Bioconductor; <<, direction of gene transcription; *, peaks with scores that are considered statistically significant (false discovery rate < 0.05). B and C, Transcriptional regulation of LAFL genes. Relative expression was determined by quantitative real-time reverse transcription-PCR (qPCR) in 35S:BBM-GR and Columbia-0 (Col-0) seeds 1 d after seed plating. Samples were treated for 3 h with dexamethasone (DEX) and cycloheximide (CHX; both at 10 µm). B, Error bars indicate se values of the three biological replicates. Statistically significant differences (asterisks) between DEX+CHX-treated 35S:BBM-GR and DEX+CHX-treated Col-0 were determined using Student’s t test (P < 0.01). C, LEC1:LEC1-GFP regulation by BBM. Samples were treated with 10 µm DEX 1 d after plating and imaged on subsequent days as indicated. The images show the adaxial sides of cotyledons, unless indicated (ab, abaxial side). The green signal in Col-0 and LEC1:LEC1-GFP cotyledon tips is autofluorescence. Bars = 250 µm.