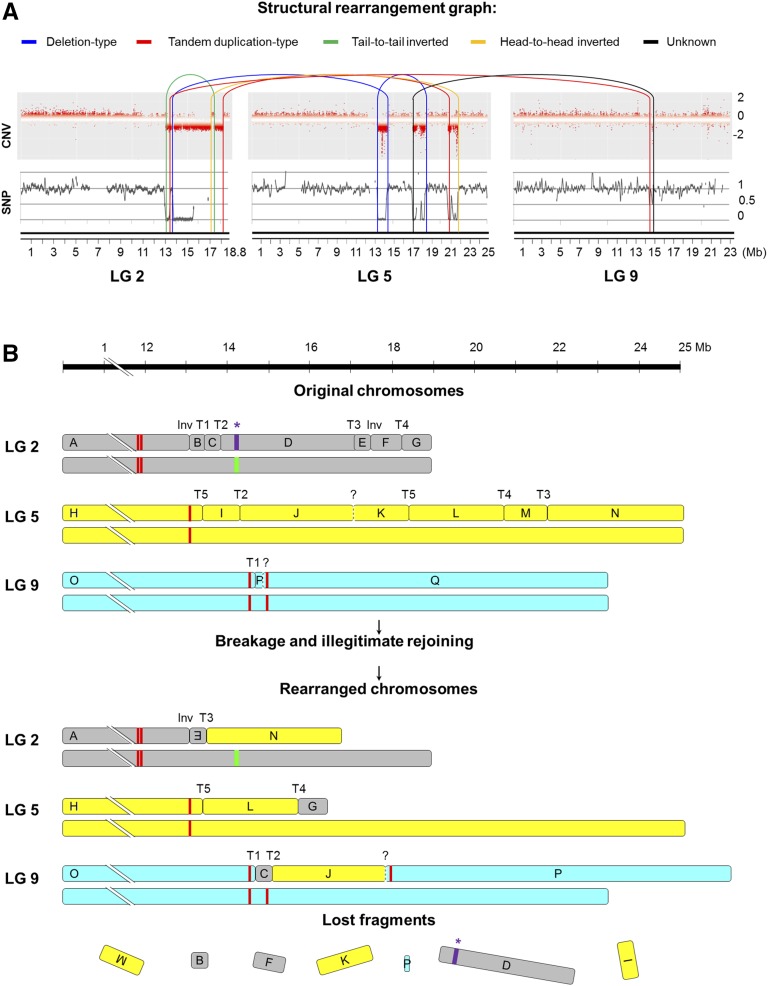
Figure 6.
DNA structural rearrangement graph and predicted rearranged chromosomes in TB. A, Structural rearrangement graph depicting CNV, LOH, and rearrangement joins and types in the affected LGs. CNV is shown as log2 (copy number ratio TB/TT) calculated for each window of the affected chromosomes by CNV-seq. SNP heterozygosity level is depicted as log2 (heterozygous SNPs per 100-kb ratio TB/TT). Connections between rearranged fragments are depicted by curved lines colored as indicated at the top of the image according to join types observed in the paired-end alignment analysis. B, Diagram depicting the original and the predicted rearranged orders of chromosome fragments affected by the chromothripsis-like restructuring that originated the variant genome of TB. Deleted fragments in TB were predicted by LOH and CNV from WGS data, and their extremes were confirmed by PCR of breakpoint junctions in most cases. Fragment order in TB derivative chromosomes was predicted considering the six deleted fragments and the six breakpoint junction sequences detected in the discordant read analysis that were validated by PCR (Inv, inversion; T, translocation). Question marks represent an additional breakpoint junction that was hypothesized from WGS read pairs that collectively did not pass all the SV analysis significance thresholds. According to this model, no other undetected breakpoint would be required to explain this catastrophic rearrangement. Rearrangements are represented in a single copy of the affected chromosomes according to the linkage of deleted fragments in the same chromosome haplotype, which was inferred from a TT self-cross progeny segregation analysis and WGS data of TB. In purple, the asterisk indicates the segment comprising the cluster of MybA genes belonging to the grape color locus functional allele, which is present in the heterozygous state in TT and is lost after genome rearrangements in TB. The white allele is represented by the green line. Red lines denote centromeric repeats according to predictions on the grapevine reference genome (Di Gaspero and Foria, 2015; Genoscope Web site).