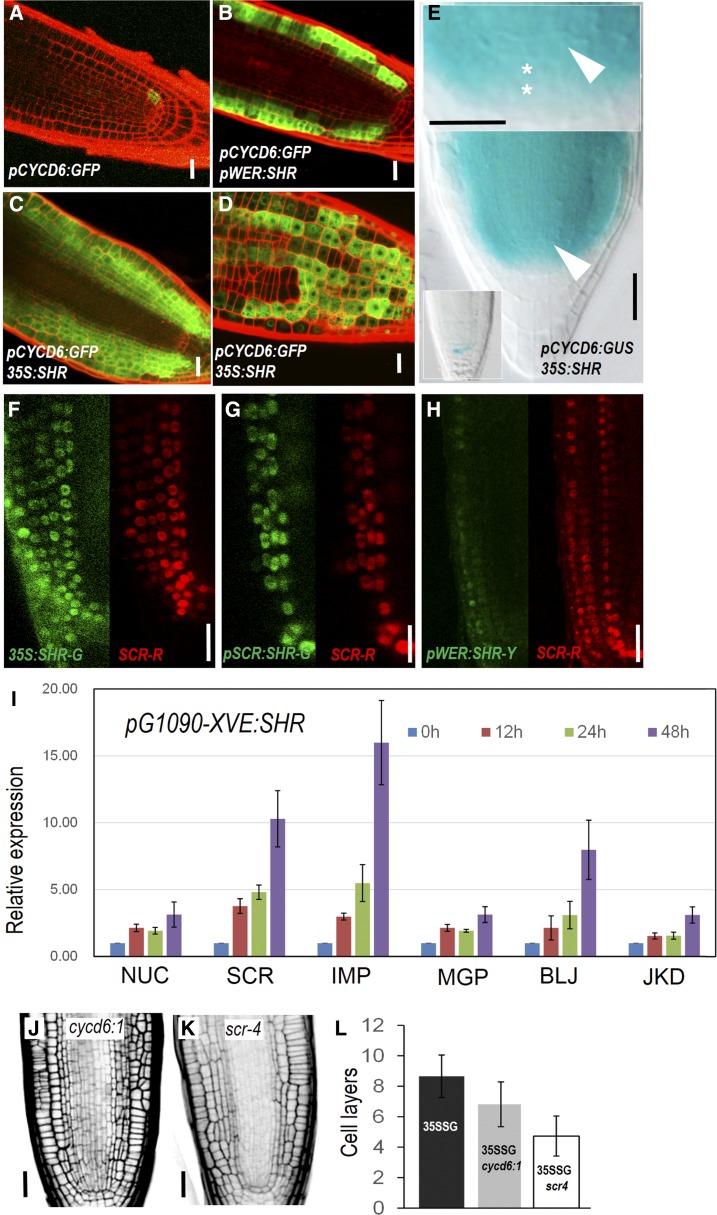
Figure 2.
SHR-mediated regulatory network is conserved across different cell types. A to D, Expression of pCYCD6;1-GFP in different backgrounds. E, pCYCD6;1-GUS is activated in the CSC region in 35S:SHR-expressing roots. White arrowheads point to the QC, and asterisks represent the CSC cells. F to H, Expression of pSCR:SCR-mCherry (SCR-R) in different backgrounds. 35S:SHR-G, 35S:SHR-GFP; pSCR:SHR-G, pSCR:SHR-GFP; pWER:SHR-Y, pWER:SHR-YFP. I, qRT-PCR analysis of SHR targets in pG1090-XVE:SHR-expressing roots at different time points. NUC, NUTCRACKER; SCR, SCARECROW; IMP, IMPERIAL EAGLE; MGP, MAGPIE; BLJ, BLUEJAY; JKD, JACKDAW. Bars show mean ± sem of three biological replicates. J and K, Radial patterning of roots expressing 35S:SHR in cycd6;1 and scr-4 mutants. L, Quantitative comparison of cell layers outside of stele. 35SSG, p35S:SHR-GFP. Data represented are mean ± sd of 14 to ∼18 roots for each sample. Scale bars = 20 μm.