Fig. 3.
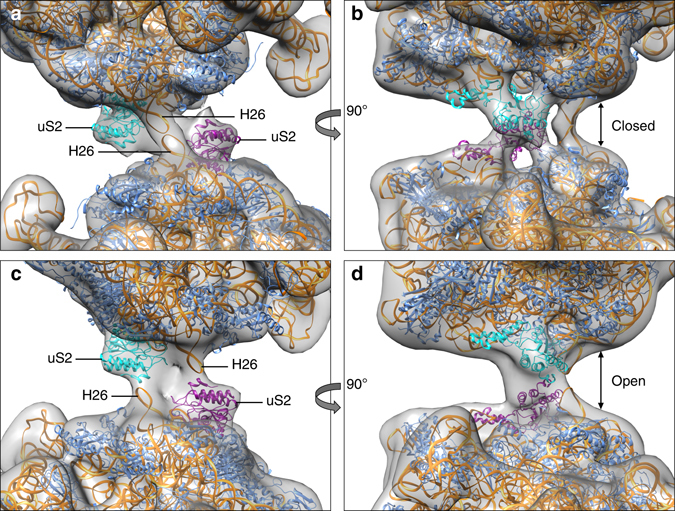
Different conformational states of 100S ribosomes. Rigid-body fits of two 70S models without the C-terminal domains of HPFlong into the low-resolution maps of the two most extreme conformations, closed state (a, b) and open state (c, d). The difference between the two classes depicts a 55° rotation around the interface. The ‘closed’ conformation closely resembles our 5.6 Å map. Colors indicate 16S rRNA (orange), protein uS2 (cyan and purple) and other proteins (blue). The rotation causes H26 to change its interaction from H26 to protein uS2 from the other ribosome, effectively widening the space between the two 70S ribosomes within the dimer (arrows in b, d)
