Fig. 6.
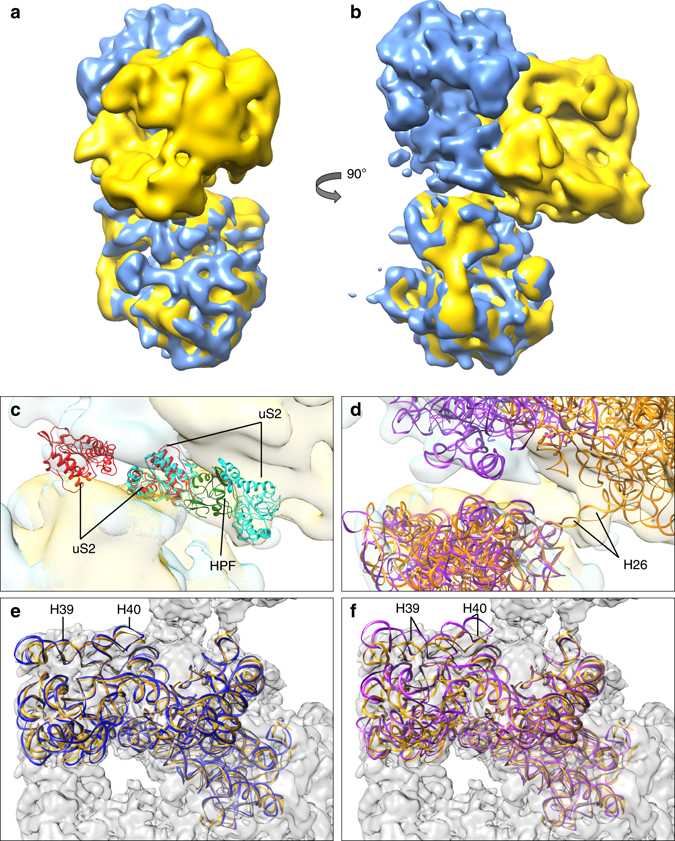
Ribosome dimerization: RMF vs. HPFlong. a–d Comparison of 3D maps of E. coli (blue 9) and L. lactis (yellow) 100S ribosomes. The density for the E. coli ribosome was obtained by taking the maximum voxel value of two copies of the EM-density map from Kato et al.9. Two copies of the L. lactis model and two copies of the T. thermophilus structure with RMF (PDB-code 4V8G), thus displaying the mechanism of E. coli, not T. thermophilus, were fitted into the corresponding density (c, d). c Zoom in of dimer interface showing proteins uS2 (marked red for T. thermophilus and cyan for L. lactis). In L. lactis the dimerization process is mediated by the C-terminal domain of HPFlong (green), whereas in T. thermophilus uS2 interacts with proteins uS3, uS4, and uS5 from the opposing ribosome (previously described as second contact site11). d The RNA chains of the L. lactis 100S (orange) and T. thermophilus 100S structures (purple). Helix 26 plays a key role in dimerization of L. lactis ribosomes but not in T. thermophilus. e, f Comparison of the 16S rRNA of ribosomes from L. lactis (orange) to T. thermophilus in apo-state (e, blue) and RMF-bound state (f, purple). The 100S map from L. lactis is depicted in gray in the background. By looking at the helices 39 and 40 of the head domain, it is clear that the ribosome from L. lactis is in the apo-state and does not undergo a conformational change upon binding of HPFlong
