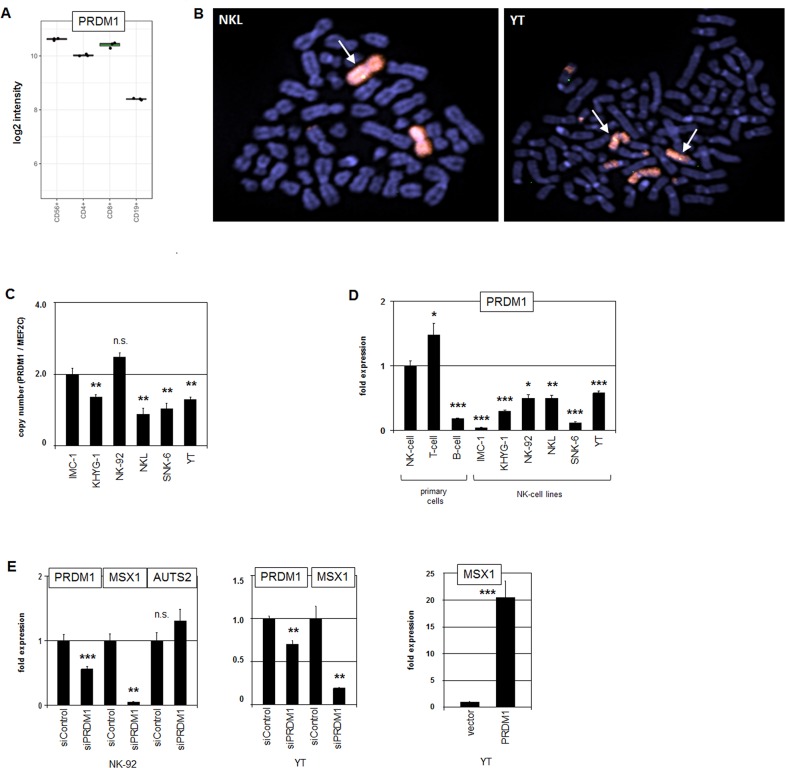
Figure 4. Analysis of PRDM1: expression, genomics and target gene regulation.
(A) Expression profiling data for PRDM1 of CD56+ NK-cells, CD4+ T-cells, CD8+ T-cells and CD19+ B-cells obtained from dataset GSE72642 are depicted in a boxplot. (B) FISH analysis of the PRDM1 locus in NKL (left) and YT cells (right). The PRDM1 probe was labelled green (appearing yellow) and the probe for the whole chromosome 6 red. PRDM1 loci are indicated by an arrow. Both cell lines exhibit a loss of one PRDM1 allele at 6q21. (C) Copy number analysis of the PRDM1 locus in NK-cell lines by RQ-PCR indicates a loss of PRDM1 in KHYG-1, NKL, SNK-6 and YT but not in IMC-1 and NK-92. The values and statistics are given in relation to IMC-1. (D) RQ-PCR analysis of PRDM1 expression in primary NK- and T-cells and six NK-cell lines revealed suppressed transcript levels in malignant NK-cells. The values and statistics are given in relation to primary NK-cells. (E) RQ-PCR analysis of PRDM1, MSX1 and AUTS2 expression after siRNA-mediated knockdown of PRDM1 in NK-92 cells (left), and RQ-PCR analysis of PRDM1 and MSX1 expression after siRNA-mediated knockdown of PRDM1 in YT cells (middle). RQ-PCR analysis of MSX1 expression after forced expression of PRDM1 in YT cells (left). The data indicate that PRDM1 activates MSX1 transcription.