Figure 1. Bioinformatics analysis shows MEOX2 and RNA Pol II occupancy throughout the lung cancer epigenome to identify a new MEOX2 gene promoter target profile.
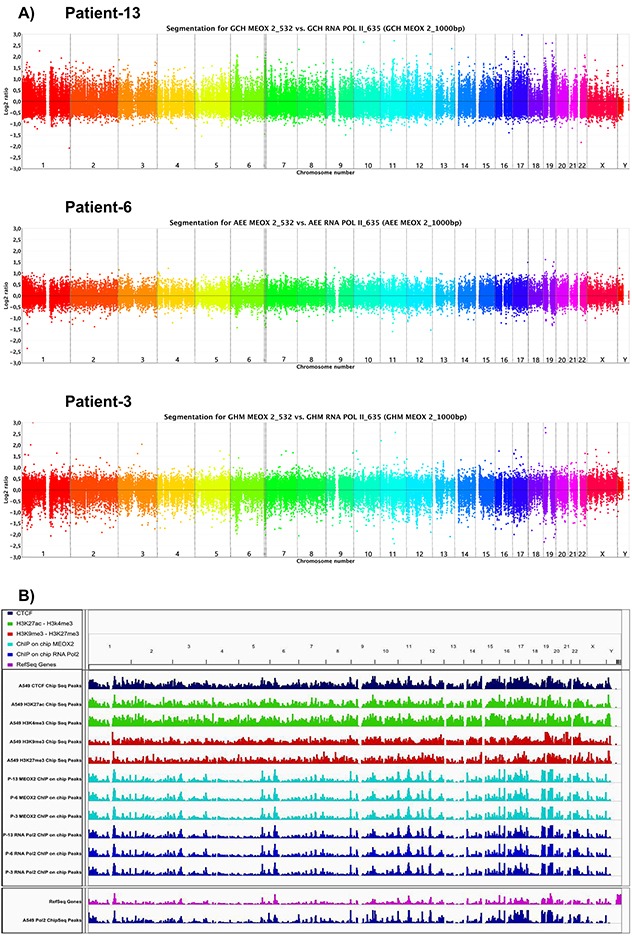
(A) Ideograms obtained via the CGH method indicate peaks of fluorescence intensity for channels 532/635, representing the enrichment of MEOX2_532 (above) and RNA Pol II_635 (bottom) throughout the lung cancer epigenome derived from lung adenocarcinoma patients. (B) Lung cancer epigenome, showing occupation of the CTCF insulator (dark-blue marks), activation histones H3K4me3 and H3K27Ac (green marks), and repression histones H3K9me3 and H3K27me3 (red marks) derived from A549 lung adenocarcinoma cells obtained from the ENCODE project database (deposited by the Broad Laboratory). Epigenome occupation by the transcription factor MEOX2 (turquoise marks) and RNA Pol II (dark-blue marks) in lung adenocarcinoma patients (generated by the present study). Additionally, the mRNA transcriptome profile (pink marks below) and RNA Pol II (dark-blue marks) epigenome occupation derived from A549 lung adenocarcinoma cells, taken from the ENCODE project database (deposited by the HudsonAlpha laboratory) are shown. All ENCODE tracks correspond to ChIPSeq assays, while present work tracks correspond to ChIP-on-chip assays utilizing gene promoter tiling arrays. All data were analyzed and visualized using IGV Viewer Program (version 2.3.60). Color codes have been included as a figure legend; increasing color intensity indicates the enrichment of epigenetic and transcriptional markers.
