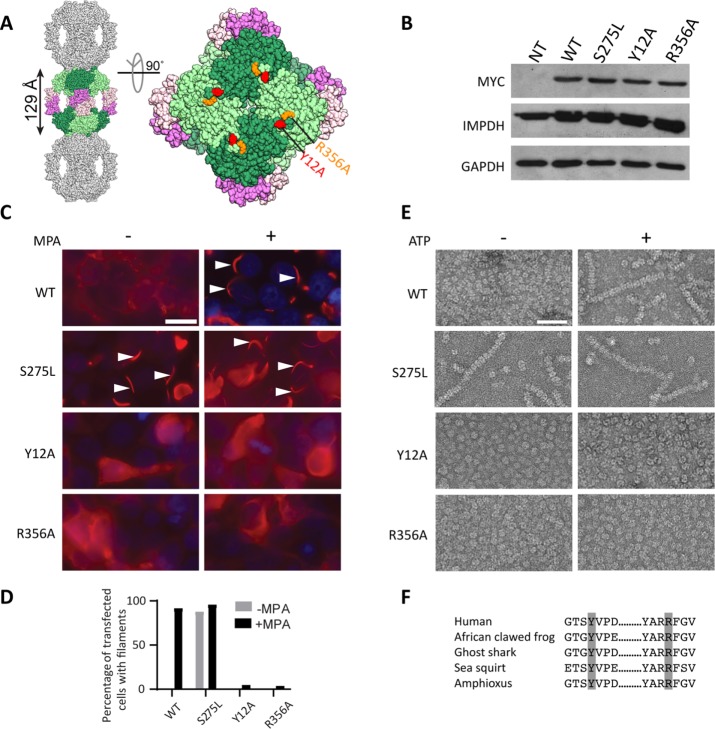
FIGURE 1:
Identification of filament disrupting and promoting IMPDH2 mutants. (A) The human IMPDH2 octamer (PDB file 1NF7). One octamer (greens, catalytic domains; pinks, Bateman domains) is shown with a subset of crystal packing neighbors (gray). Tyrosine 12 (red) and arginine 356 (orange) are indicated. (B) Immunoblot of HEK293 cell lysates with antibodies against myc-tagged IMPDH2, total IMPDH, or loading control glyceraldehyde 3-phosphate dehydrogenase (GAPDH). NT, not transfected. (C) Anti-myc immunofluorescence of HEK293 cells transfected with the indicated IMPDH2-myc constructs (red). 4’6-diamidino-2-phenylindole (DAPI) staining in blue. Cells were either treated with 10 µM mycophenolic acid (MPA) to induce IMPDH2 filaments (arrowheads) or not, as indicated. Images are representative of three experiments. Scale bar, 20 µm. (D) Quantification of transfected cells with filaments; n = 100 cells per sample. (E) Negative-stain electron microscopy of purified IMPDH2 proteins incubated with 5 mM NAD+, with and without 1 mM ATP. Images represent two separate experiments. Scale bar, 50 nm. (F) Evolutionary conservation of tyrosine 12 and arginine 356. IMPDH amino acid sequences from Homo sapiens, Xenopus laevis, Callorhinchus milii, Ciona savignyi, and Branchiostoma belcheri were retrieved and aligned using Consurf (Ashkenazy et al., 2016).