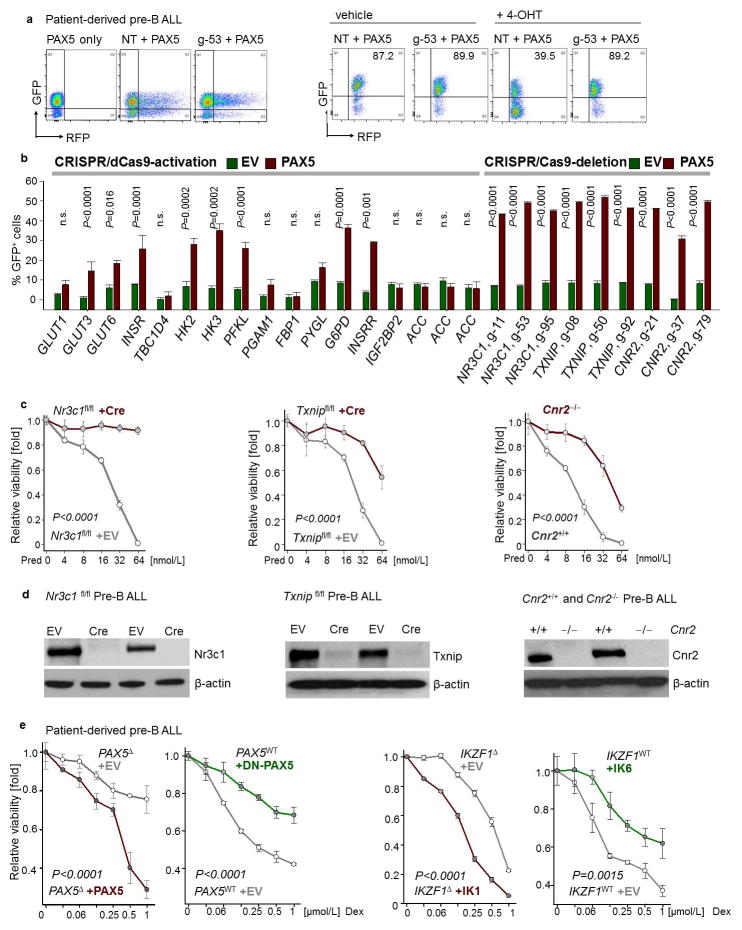
Extended Data Figure 9. Mechanistic contribution of PAX5 targets.
a, b, Representative FACS plots from CRISPR-based gene editing experiments (a). CRISPR complexes were delivered to patient-derived PAX5-haploinsufficient pre-B ALL cells along with RFP-tagged gRNAs to direct dCas9 (CRISPR-mediated gene activation) or Cas9 (CRISPR-mediated gene deletion) to specific PAX5 target genes (e.g. NR3C1; left). Upon inducible activation of GFP-tagged PAX5 or EV in patient-derived pre-B ALL cells, enrichment or depletion of GFP+ cells carrying RFP-tagged gRNAs (RFP+) was monitored by flow cytometry (right; NT: non-targeting; g-53: gRNA clone 53 for deletion of NR3C1). Changes in percentage of GFP+ cells carrying the indicated gRNAs following induction, as compared to cells carrying the NT gRNA (b). c, d, In murine pre-B ALL models for genetic loss of Nr3c1, Cnr2, and Txnip function, responses to prednisolone (Pred) were measured (c) and protein levels of Nr3c1, Txnip and Cnr2 were examined (d). (e) In a patient-derived PAX5-haploinsufficient pre-B ALL (PAX5Δ; left), Dex responses upon inducible activation of PAX5 or empty vector (EV) were measured. In a patient-derived PAX5-wildtype pre-B ALL (PAX5WT; right), effects of DN-PAX5 on Dex responses were measured. Likewise, dose-response curves for Dex were measured in two patient-derived pre-B ALL samples carrying either wildtype or deleted IKZF1 when DN-IKZF1 or IKZF1 were inducibly expressed, respectively. Data shown as mean from 3 independent experiments (± s.d.) and assessed by two-tailed t-test (b) or two-way ANOVA (c, e).