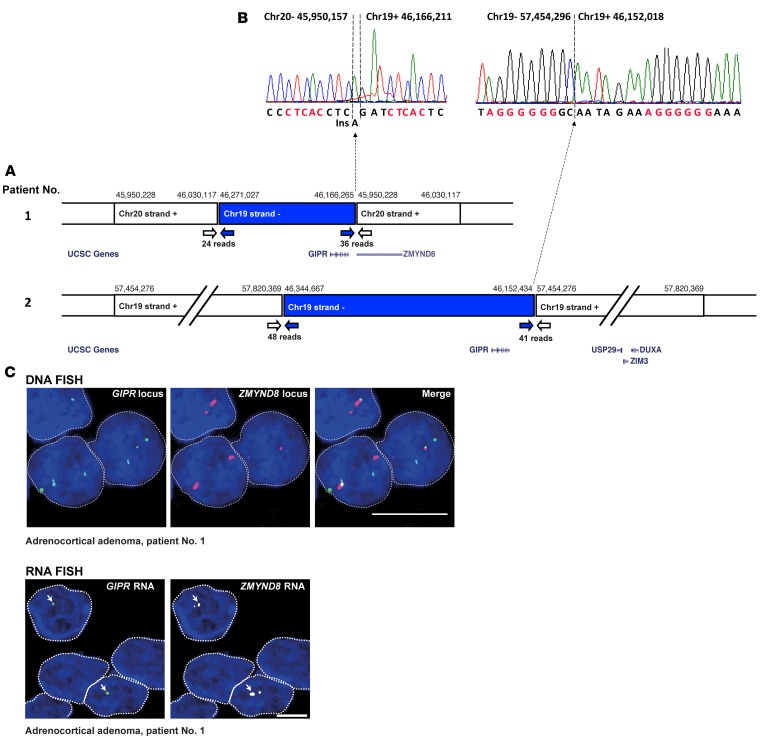
Figure 3. Chromosome rearrangements in adrenocortical adenoma of two patients with glucose-dependent insulinotropic polypeptide–dependent (GIP-dependent) Cushing’s syndrome drive ectopic GIP receptor (GIPR) expression in adenoma cells.
(A) Genomic rearrangements in adenoma samples from patient #1 and #2, as revealed by mate-pair analysis. Next-generation sequencing (NGS) data identified abnormally mapped reads corresponding in tumor from patient #1 to interchromosomal rearrangement between chromosome segments 19q13.32 and 20q13.12, and in tumor from patient #2 to intrachromosomal rearrangement between chromosome segments 19q13.32 and 19q13.43. The genomic coordinates of the rearranged genomic segments are shown above the boxes. White and blue arrows indicate abnormal reads corresponding to break points. Selected genes in the rearranged chromosome segments are shown below the boxes (adapted from the UCSC Genome Browser). The break point at position hg19 chr20:45,950,228 in adenoma from patient #1 corresponds to the second intron of the zinc finger and the MYND (myeloid, Nervy, and DEAF-1) domain containing 8 (ZMYND8) gene, which is consequently truncated. In the adenoma of patient #2, the break point at position hg19 chr19:57,454,276 falls within a noncoding region of chromosome 19. (B) The breakpoints in the proximity of the GIPR gene at base-pair resolution, characterized by conventional Sanger sequencing of the PCR fragments, showing one base-pair insertion (Ins) typical of canonical nonhomologous end-joining. Microhomologies are indicated in red. (C) DNA (top) and RNA FISH (bottom) analyses in adenoma of patient #1. On DNA FISH, 3 GIPR loci (green) and two ZMYND8 loci (red) are visible in each interphase nucleus. Thus, DNA FISH confirms duplication of the 19q13.32 region and its insertion in close proximity to ZMYND8 in this sample. Representative examples of GIPR expression (green) and of ZMYND8 expression (white) in adenoma of patient #1, as analyzed by RNA FISH, are also shown. Arrows indicate GIPR expression from a single allele in each nucleus. The RNA FISH signal corresponds to the translocated GIPR allele fused with intron 2 of the ZMYND8 gene located in chromosome region 20q13.12. Scale bars: 5 μm.