Abstract
Heart disease remains the leading cause of death worldwide, highlighting a pressing need to identify novel regulators of cardiomyocyte (CM) function that could be therapeutically targeted. The mammalian Hippo/Tead pathway is critical in embryonic cardiac development and perinatal CM proliferation. However, the requirement of Tead1, the transcriptional effector of this pathway, in the adult heart is unknown. Here, we show that tamoxifen-inducible adult CM–specific Tead1 ablation led to lethal acute-onset dilated cardiomyopathy, associated with impairment in excitation-contraction coupling. Mechanistically, we demonstrate Tead1 is a cell-autonomous, direct transcriptional activator of SERCA2a and SR-associated protein phosphatase 1 regulatory subunit, Inhibitor-1 (I-1). Thus, Tead1 deletion led to a decrease in SERCA2a and I-1 transcripts and protein, with a consequent increase in PP1-activity, resulting in accumulation of dephosphorylated phospholamban (Pln) and decreased SERCA2a activity. Global transcriptomal analysis in Tead1-deleted hearts revealed significant changes in mitochondrial and sarcomere-related pathways. Additional studies demonstrated there was a trend for correlation between protein levels of TEAD1 and I-1, and phosphorylation of PLN, in human nonfailing and failing hearts. Furthermore, TEAD1 activity was required to maintain PLN phosphorylation and expression of SERCA2a and I-1 in human induced pluripotent stem cell–derived (iPS-derived) CMs. To our knowledge, taken together, this demonstrates a nonredundant, novel role of Tead1 in maintaining normal adult heart function.
Keywords: Cardiology, Cell Biology
Keywords: Calcium, Heart failure, Transcription
Tead1 deletion causes acute dilated cardiomyopathy in mice and is required to maintain SERCA2a expression.
Introduction
Heart failure is a global health problem affecting more than 25 million people worldwide, including 5.7 million adults in the United States (1). Dilated cardiomyopathy (DCM) — a form of heart failure with diminished cardiac contractility, dilated ventricular chambers, myocyte death, and myocardial fibrosis (2) — has a has a prevalence of 1 in 2,500 (3, 4) and a high 5-year mortality of 50% (4). DCM may result from defects in diverse cellular processes, including those resulting from ischemia/injury and familial genetic defects. Familial DCM — involving more than 40 genes encoding nuclear envelop proteins, sarcomeric proteins, gene transcription and splicing machinery, and Ca2+-handling proteins — accounts for up to half of nonischemic DCM (5). However, in the majority of cases, the cause of DCM remains idiopathic and the underlying regulatory pathways are still ill defined.
The mammalian Hippo/Tead pathway is an evolutionarily conserved pathway that regulates cell proliferation, differentiation, and organ size in many tissues and has been implicated in many forms of cancer (6, 7). Recent studies demonstrate the critical role of the mammalian Hippo/Tead pathway in cardiac development, cardiomyocyte (CM) proliferation, and regeneration (8–12). TEA domain family members Tead1-4, the downstream transcriptional effectors of the Hippo pathway, confer transcription activity via binding predominantly SV40 GT-IIC (5′-GGAATG-3′) and Muscle-CAT (5′-CATTCCT-3′) (13) elements and play critical roles in cell proliferation, lineage differentiation, and tumor formation (11, 14, 15). Of the 4 members of the TEAD family, Tead1 is the most abundant in the heart and plays a nonredundant role in cardiac development, as germline deletion of Tead1 led to cardiac hypoplasia and embryonic lethality at E11.5 (16). Yap and Taz, coactivators of Tead1, are critical for CM proliferation in perinatal cardiac development (14), while Vgll4, a corepressor of Tead1, regulates CM gene expression and inhibits early postnatal heart growth (17, 18). Presence of Tead-binding enhancers in multiple genes critical for cardiac function (19–21) and continued expression of Tead1 protein in the postnatal heart led us to hypothesize that Tead1 plays a broader transcriptional regulatory role in postproliferative CMs and is required for normal adult cardiac function.
To test this hypothesis, we generated an inducible CM-specific Tead1 conditional KO mouse model. Tead1 deletion, specifically in adult murine CMs, resulted in acute-onset lethal DCM with profound excitation-contraction coupling impairment, associated with loss of sarcoplasmic/ER Ca2+ ATPase-2a (SERCA2a) activity and markedly reduced sarcoplasmic reticulum (SR) Ca2+ load. Mechanistically, Tead1 directly enhanced SERCA2a and SR-associated protein phosphatase 1 regulatory subunit, Inhibitor-1 (I-1), expression. Thus, Tead1 maintains SERCA2a activity by enhancing the phosphorylation of phospholamban (Pln) via inhibition of SR-associated protein phosphatase 1 (PP1) activity. Additionally, not only is there a trend for correlation between protein levels of TEAD1, I-1, and phosphorylation of PLN in human nonfailing and failing hearts, but in addition, blocking TEAD1 activity in human induced pluripotent stem cell–derived (iPS-derived) CMs revealed that TEAD1 is essential in maintaining PLN phosphorylation and expression of SERCA2a and I-1, strongly suggesting that this regulatory pathway is also conserved in human CMs.
Results
Tead1 continues to be expressed in adult heart.
The mammalian Hippo-Tead1 pathway plays a critical role in perinatal CM proliferation (22, 23). Tead1 is robustly expressed in embryonic and early postnatal heart, periods of high CM proliferation (24). However, Tead1 continues to be expressed in the adult heart, albeit at a lower level (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.93343DS1). Recent studies on its corepressor, Vgll4, in early postnatal hearts indicate that decreased function of Tead1 affects postnatal heart growth. However, the specific function of Tead1 in the adult heart function remains an unanswered question. To address this fundamental question, we generated mice with inducible Tead1 deletion only in adult mouse CM to study the nonredundant role of Tead1 in adult heart function.
Tead1 deletion in adult CM leads to severe acute-onset heart failure with a DCM phenotype.
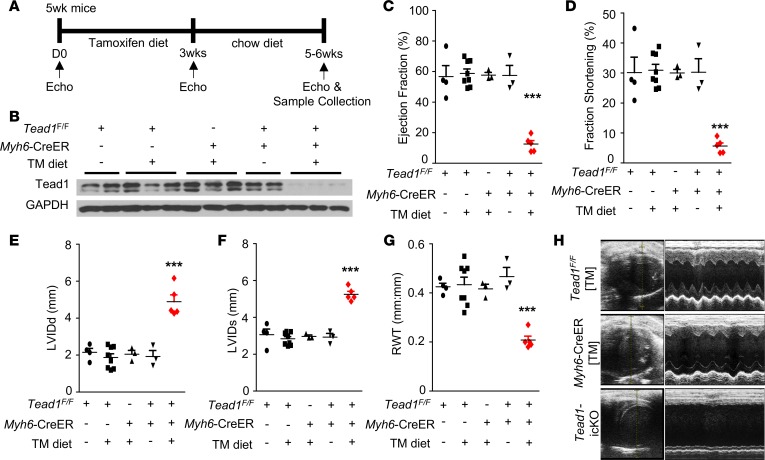
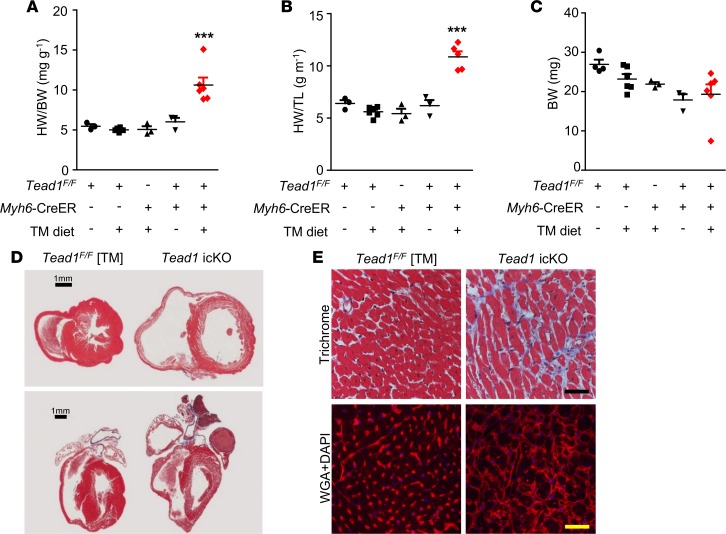
To delete Tead1 specifically in CM in a temporal and spatially inducible manner, we first generated mice carrying Tead1 alleles, wherein exon 3 (encoding the DNA binding domain) was flanked by loxP sites (Tead1F/F) (Supplemental Figure 2A and Supplemental Methods). We then crossed these Tead1-floxed mice with Myh6-MerCreMer (indicated herein as Myh6-CreER) deleter mice (25) to achieve tamoxifen-inducible (TM-inducible) CM-specific Tead1 deletion (indicated herein as Tead1-icKO) (Supplemental Figure 2, A–D). Since high doses of TM, administered as i.p. bolus injections, have been reported to induce myocardial dysfunction in mice carrying the Myh6-CreER allele (26, 27), we adopted a 3-week TM diet protocol (28) followed by regular chow in 5-week-old mice to achieve CM-specific deletion of Tead1 (Figure 1A). This protocol resulted in efficient Tead1 deletion in CM (Figure 1B), but not in skeletal muscle (Supplemental Figure 2E) or other tissues (data not shown). Death occurred starting on the fifth week of the protocol and Tead1-icKO mice displayed almost 100% mortality by the end of the sixth week. Echocardiography, at 3 weeks after initiating TM diet, demonstrated significant worsening in cardiac function, accompanied by ventricular dilatation and impaired LV contractility in Tead1-icKO mice (Supplemental Figure 3). By 6 weeks, ejection fraction (EF) (12.6% ± 5.0% vs. 57.9% ± 9.0% in controls) and fractional shortening (FS) (6.2% ± 2.3% vs. 30.5% ± 6.3% in controls) were profoundly diminished (Figure 1, C–D), along with chamber dilatation (Figure 1, E–H, and Supplemental Videos 1 and 2). This was accompanied by increased heart weight of Tead1-icKO mice (normalized either to body weight or to tibial length), without a significant change in body weight (Figure 2, A–C). All of this was consistent with a rapid-onset cardiac insufficiency. Histopathology confirmed thin-walled biventricular and atrial dilatation with occasional left atrial fibrin clots (Figure 2D). Trichrome, wheat germ agglutinin (WGA) (Figure 2E), and TUNEL (Supplemental Figure 4) staining demonstrated that there was accompanying interstitial fibrosis with disruption of normal myocardial architecture, without a significant change in apoptosis. All this, together, was consistent with severe DCM being induced with CM-specific deletion of Tead1.
Figure 1. Tead1 ablation in adult cardiomyocytes leads to acute diminished cardiac contractility.
(A) Experimental procedure and tamoxifen diet protocol for inducing CM-specific Tead1 KO (Tead1-icKO). (B) Tead1 protein expression in the heart ventricles. GAPDH, loading control. TM, tamoxifen. (C) Ejection fraction, (D) fractional shortening, (E) left ventricular internal diameter at end diastole (LVIDd), (F) left ventricular internal diameter at end systole (LVIDs), and (G) relative wall thickness (RWT) by echocardiography (Echo) at 6 weeks after starting TM diet. Values are shown as mean ± SEM. ***P < 0.001; data were analyzed by 1-way ANOVA with Tukey’s post hoc test. Comparisons between Tead1-icKO mice (n = 6–8) and TeadF/F chow diet mice (n = 4), TeadF/F TM diet mice (n = 8), Myh6-CreER TM diet mice (n = 3), or TeadF/F;Myh6-CreER chow diet mice (n = 3) were statistically significant. No significance was present among the four control groups. (H) Representative 2-D M-mode images of the left ventricle from Echo assessment of the TeadF/F TM diet, Myh6-CreER TM diet, and Tead1-icKO mice at 6 weeks after starting TM diet.
Figure 2. Tead1 deletion in adult cardiomyocytes leads to acute dilated cardiomyopathy.
(A) heart weight/body weight (HW/BW) ratio, (B) heart weight/tibial length (HW/TL) ratio, and (C) body weight, all after 6 weeks of the diet protocol. ***P < 0.001; data were analyzed by 1-way ANOVA with Tukey’s post hoc test. Comparisons between Tead1-icKO mice (n = 6) and TeadF/F chow diet mice (n = 4), TeadF/F TM diet mice (n = 4), Myh6-CreER TM diet mice (n = 3), or TeadF/F;Myh6-CreER chow diet mice (n = 3). No significance was present among the 4 control groups. (D) Trichrome staining of transverse and coronal section of the hearts from the TeadF/F TM diet and Tead1-icKO mouse. Scale bars: 1 mm. (E) Trichrome staining and wheat germ agglutinin (WGA, red; DAPI, blue) staining. Scale bars: 40 μm.
Tead1 is required for normal heart function in older mice.
The above results demonstrate the requirement for Tead1 in younger adult (age 5–12 weeks old) mouse CM function. Since there was an age-dependent reduction in Tead1 protein (Supplemental Figure 1) in the heart, we asked whether Tead1 continues to have a critical function in CM, even when expressed at much lower levels in older mice. To test this, we deleted Tead1 specifically in CM of older mice using the TM diet protocol described above. Older mice (16 weeks old) were started on the TM diet for 3 weeks and were then shifted to regular chow and assessed at age 21–22 weeks. Similar to the younger mice with Tead1-icKO, Tead1 deletion in older mice also led to profound heart failure, with echocardiographic evidence of a significant decrease in EF and FS accompanied by profound chamber dilatation (Supplemental Figure 5, A and B). These Tead1-deleted hearts weighed more and displayed a disruption of myocardial architecture on histology (Supplemental Figure 5, C and D). The similar course of rapid-onset DCM in both younger and older mice with inducible Tead1-icKO demonstrated the continued requirement of Tead1 in adult heart function.
Tead1 is required for normal SERCA2a expression and activity to maintain CM SR-cytosolic calcium cycling.
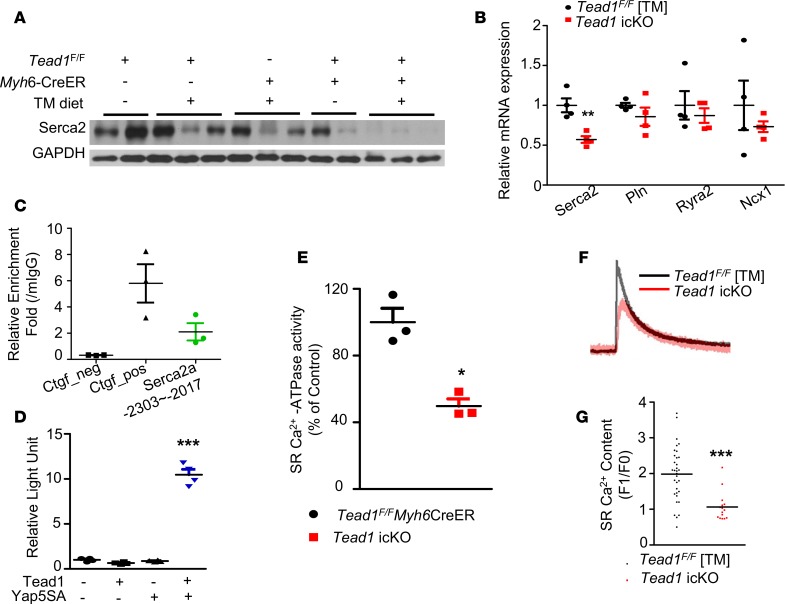
Disruption of excitation-contraction coupling is a possible mechanism for rapid-onset heart failure. SERCA2a is the primary mediator of calcium reuptake into the SR, a critical component of excitation-contraction coupling (29). A decrease in SERCA2a expression and activity, leading to a disruption of normal calcium cycling across the SR and consequent impairment of excitation-contraction coupling, is a frequent common denominator in heart failure (30). Serca2/Atp2a2 transcript and protein levels were significantly lower in Tead1-icKO hearts (Figure 3, A and B). Since Serca2a expression may be decreased in end-stage heart failure of different etiologies, we assessed Serca2a protein level in the early stages (within a week after discontinuing TM diet) after CM-Tead1 deletion and found it significantly lower than the controls (Supplemental Figure 6). This suggested that the decrease in the Serca2a level may be an underlying initiating mechanism for heart failure in Tead1-deleted hearts. We then tested if Serca2a was directly regulated by Tead1. Serca2a promoter harbors multiple Tead-binding motifs (Supplemental Table 1), and we confirmed Tead1 occupancy by ChIP assay, suggesting direct transcriptional regulation of Serca2a by Tead1 in vivo (Figure 3C). We confirmed that this regulation of Serca2a promoter by Tead1 was functional, using the native Serca2a promoter luciferase reporter assay with overexpression of Tead1 alone or coexpressed with its constitutive active coactivator, Yap5SA, in H9c2 cardiomyoblast cell line. We showed that Tead1 alone (which requires a coactivator for transcriptional activity) does not change the Serca2a promoter activity, but when coexpressed with its coactivator Yap5SA, it was sufficient to increase Serca2a promoter activity (Figure 3D), indicating that Tead1-mediated transcriptional regulation of Serca2a is direct and cell-autonomous. Further experimentation revealed that, not only was the level of Serca2a decreased, but the SR-associated SERCA activity (normalized to mg protein and expressed as a percentage of control hearts) was also decreased significantly by 46.3% (Figure 3E). A decrease in SERCA activity would be expected to lead to a decrease in SR calcium load. Indeed, CMs, isolated from Tead1-icKO ventricles, displayed a significant decrease in the amplitude of caffeine-stimulated calcium release from the SR, reflecting a decrease in SR load (F1/F0 2.13 ± 0.20 vs. 3.97 ± 0.26 in controls), when compared with controls (Figure 3, F and G).
Figure 3. Tead1 is required for maintaining cardiomyocyte SERCA2a activity.
(A) Serca2 protein level in control and Tead1-icKO hearts. (B) mRNA expression of genes related to Ca2+ cycling (n = 4). (C) ChIP from 12-week-old mouse hearts (n = 3) was performed with control mouse IgG (mIgG) and TEAD1 antibody. The presence of mSerca2a promoter was detected by qPCR. (D) mSerca2a promoter luciferase assay in H9c2 rat cardiomyoblasts. (E) SR-associated Ca2+-ATPase activity. (F) Representative Ca2+ tracing following 10 mmol/l caffeine stimulus of SR Ca2+ release. (G) Quantification of SR Ca2+ content (F1/F0). (n = 3, cell number = 15–17). ***P < 0.001, **P < 0.01, *P < 0.05; P values were determined using Student’s t test.
These findings indicate that impaired SR Ca2+ load and consequent impairment in SR calcium cycling — due to a decrease in Serca2a level, along with a decrease in SERCA activity in Tead1-deficient CMs — could be a principal mechanism underlying the reduced contractility in Tead1 CM-ablated mice.
Tead1 regulates SERCA2a activity in a cell-autonomous manner via modulating phosphorylation of Pln by transcriptional activation of I-1.
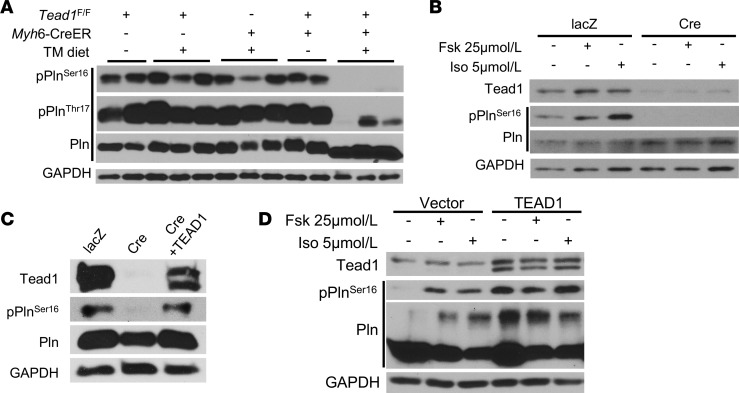
We then investigated the mechanism of the decreased SERCA activity in CM with Tead1 deletion. SERCA2a activity is allosterically inhibited by Pln. Phosphorylation of Pln at Ser16/Thr17 sites — by PKA and CaMK, respectively — relieves this inhibition and enhances SERCA2a activity (31, 32). Hence, we next measured the phosphorylation status of Pln in Tead1-deleted hearts. Consistent with the observed decrease in SERCA activity, Pln is strikingly dephosphorylated (Ser16/Thr17) in Tead1-icKO ventricles (Figure 4A).
Figure 4. Tead1 is required for the phosphorylation of phospholamban (Pln).
(A) Phosphorylation levels of Pln on Ser16/Thr17 in control and Tead1-icKO hearts. (B) Effects of foskolin (Fsk) 25 μmol/l and isoproterenol (Iso) 5 μmol/l on pPlnSer16 protein levels in lacZ or Cre adenovirus–treated Tead1-flox neonatal CMs (NCMs). (C) Immunoblot showing the rescue of phosphorylation of Pln at Ser16 by reexpression of TEAD1 in Tead1-icKO NCMs. (D) Effects of Fsk and Iso on pPlnSer16 protein levels in control vector and TEAD1-overexpressed NCMs.
Interestingly, severe heart failure, itself, has been associated with a decrease in Pln phosphorylation, raising the question of whether the observed decrease in Pln phosphorylation in Tead1-deleted hearts was due to the loss of cell-autonomous Tead1 function in CM or a nonspecific non–cell-autonomous effect of heart failure itself. To test this, we used an ex vivo model of Tead1 deletion in CM, wherein neonatal CMs (NCMs) were harvested from Tead1F/F mice and treated ex vivo with Ad-Cre (or with Ad-lacZ as control) to generate Tead1-deleted NCMs, to exclude non–cell-autonomous effects on Pln phosphorylation.
Consistent with in vivo findings, we found that ex vivo Tead1 deletion in isolated NCMs led to a marked decrease in phospho-Ser16-Pln, under basal and stimulated (forskolin or isoproterenol) conditions (Figure 4B). Restoring Tead1 levels was sufficient to rescue phospho-Ser16-Pln levels (Figure 4C). In contrast, overexpression of Tead1 in NCMs increased phospho-Ser16-Pln (Figure 4D), under both basal and stimulated conditions. These results demonstrate that Tead1 regulates the phosphorylation status of Pln in a cell-autonomous manner.
Tead1 is a cell-autonomous, direct transcriptional enhancer of I-1 and regulates dephosphorylation of Pln.
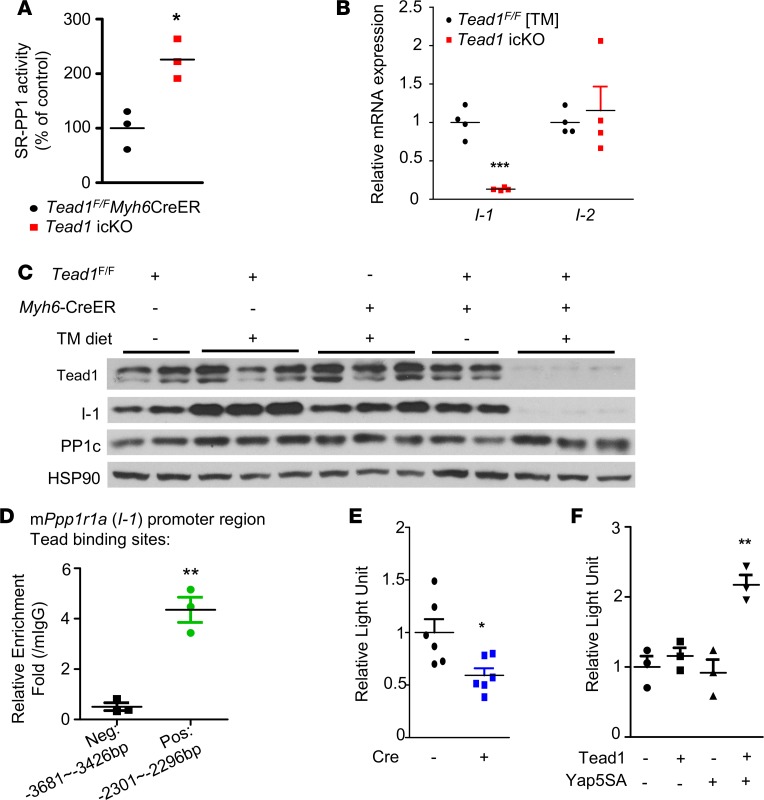
The decreased phospho-Ser16-Pln observed in Tead1-icKO hearts could be secondary to either a decrease in kinase (PKA) or an increase in phosphatase (SR-associated PP1) activity or both. PKA activity in Tead1-icKO hearts was not different from control hearts (Supplemental Figure 7); however, the SR-associated PP1 activity, which dephosphorylates phospho-Pln at Ser16 (33), was significantly higher in the Tead1-icKO hearts (Figure 5A), strongly suggesting that the decrease in phospho-Ser16-Pln in Tead1-icKO hearts was a result of increased dephosphorylation of phosho-Pln at Ser16.
Figure 5. Increased SR-associated protein phosphatase 1 (PP1) activity in Tead1-icKO hearts.
(A) SR-associated protein phosphatase 1 (PP1) activity in control and Tead1-icKO hearts (n = 3). (B) I-1 and I-2 mRNA expression levels in TeadF/F TM diet and Tead1-icKO hearts (n = 4). (C) I-1 and PP1 catalytic unit (PP1c) protein levels in control and Tead1-icKO hearts. (D) ChIP with control mouse IgG (mIgG) and TEAD1 antibody (n = 3). The presence of mPpp1r1a (I-1) promoter regions were detected by qPCR. I-1 promoter luciferase assay in (E) Tead1-KO (Cre-AdV) and control (lacZ-AdV) Tead1-flox NCMs and (F) H9c2 rat cardiomyoblasts. ***P < 0.001, **P < 0.01, *P < 0.05; P values were determined using Student’s t test.
Since the protein level of PP1-catalytic subunit (PP1c) was not decreased (Figure 5C), to explain the increased PP1-activity, we hypothesized that the inhibitors of PP1 may be downregulated with Tead1 deletion. SR-associated PP1 activity is regulated by regulatory subunits I-1 (Ppp1r1a) and I-2 (Ppp1r2) (34). I-1 transcript and protein, but not I-2, was decreased in Tead1-icKO hearts (Figure 5, B and C). I-1 promoter harbors multiple conserved Tead1 binding motifs (Supplemental Table 2), and using ChIP assay, we demonstrated Tead1 occupancy (Figure 5D). This suggested that Tead1 could be a direct transcriptional regulator of I-1. To demonstrate functional direct regulation of I-1 by Tead1, we performed a promoter luciferase reporter assay in NCMs with ex vivo Tead1 deletion using the native promoter of I-1 containing the Tead1 binding elements. This demonstrated a significant decrease (by 41%) in I-1 promoter activity with Tead1 deletion (Figure 5E), indicating Tead1 is required for normal I-1 transcription. We then confirmed the regulation of I-1 promoter by Tead1 in the H9c2 cardiomyoblast cell line. As expected, when coexpressed with its coactivator Yap5SA, Tead1 was sufficient to increase I-1 promoter activity (Figure 5F), indicating that Tead1-mediated transcriptional regulation of I-1 is direct and cell-autonomous. These data demonstrate that Tead1 has a key regulatory role in modulating SR calcium cycling and CM excitation-contraction coupling by maintaining the phosphorylated state of Pln and consequently limiting the Pln-mediated inhibition of SERCA2a pump activity under basal and adrenergic-stimulated conditions.
Tead1 deletion leads to changes in pathways regulating mitochondria and sarcomeric function.
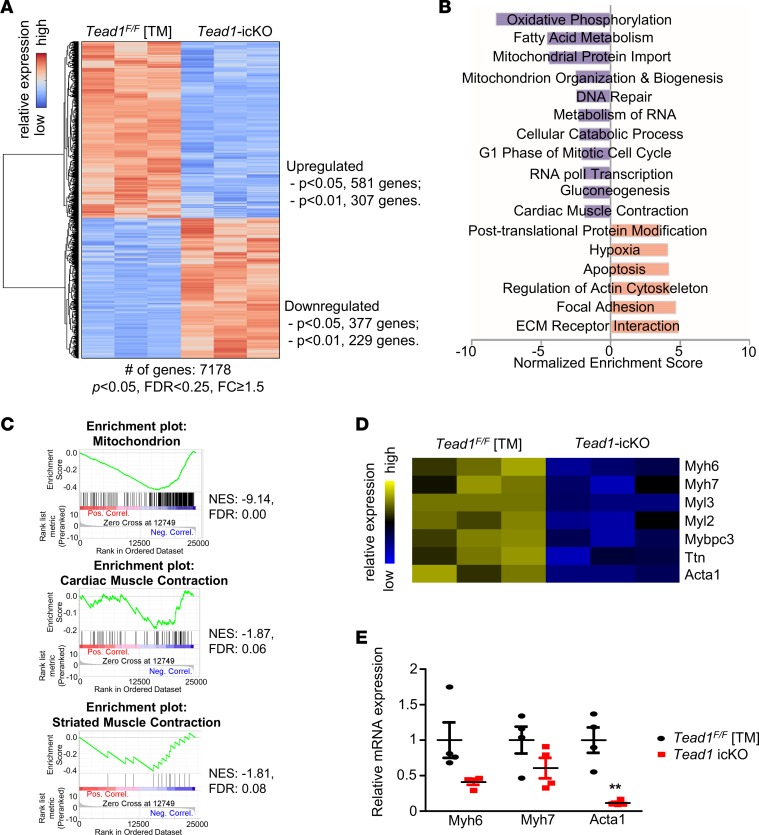
Since Tead1 is a transcription factor that is important in cardiac development and has been suggested to be a part of a cardiac-specific gene enhancer cluster (20), we explored further whether Tead1 regulates other critical pathways in adult CM (ACM). To define these pathways, we performed a global transcriptome analysis comparing Tead1-deficient and control hearts. Tead1 deletion led to an upregulation of 307 and a downregulation of 229 genes as compared with controls (Figure 6A and Supplemental Table 4). A Gene Set Enrichment Analysis (GSEA) revealed that pathways regulating mitochondrial function, cellular proliferation, and cardiac muscle contraction, among others, were significantly enriched and were downregulated with Tead1 deletion (Figure 6, B and C). Many of the genes encoding sarcomeric proteins were downregulated in the Tead1-deleted hearts and were confirmed on quantitative PCR (qPCR) (Figure 6, D and E). These strongly suggested that Tead1 plays a broader role in maintaining ACM function, in addition to specifically being required for normal excitation-contraction coupling by the regulation of SERCA2a expression and activity.
Figure 6. Global transcriptome analysis of Tead1-icKO heart.
(A) Gene clustering by Z-score using Euclidean distance metric in control (Tead1F/F TM) and icKO samples. Gene expression fold change (FC) was calculated by icKO over control. P < 0.05, FDR < 0.25, FC ≥ 1.5. (B) Selected Gene Set Enrichment Analysis (GSEA) pathways in Tead1-icKO. (C) Enrichment plots showing the selected enriched pathways in Tead1-icKO dataset. (D) Heatmap of genes encoding key structural proteins of sarcomere. (E) qPCR confirmation of mRNA expression of selected genes encoding sarcomere-related proteins differentially changed on microarray (n = 4).
TEAD1 regulates PLN phosphorylation and I-1 in human CMs.
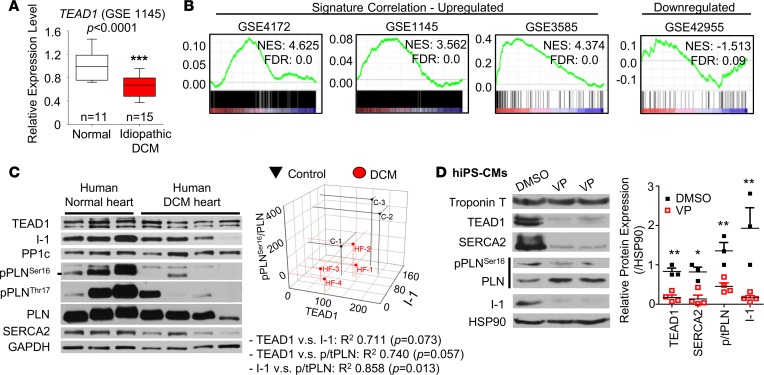
Whether TEAD1 is similarly required for normal excitation-contraction coupling in human CM is unknown. Data from GEO datasets revealed that Tead1 expression was decreased in the ventricles of human DCM patients (Figure 7A). Consistent with this, GSEA analysis in Tead1-icKO heart microarray dataset (GSE85038, see Supplemental Methods), revealed a significant enrichment of both upregulated and downregulated gene sets from previously reported human DCM gene signatures (Figure 7B), suggesting that similar pathways may be regulated. TEAD1 protein tends to be decreased (~20% decrease as compared with nonfailing control hearts) in human DCM hearts in our data, though not reaching to the levels of statistical significance. However, its expression displayed a positive correlation with SERCA2a, I-1, and PLN phosphorylation levels (Figure 7C) — congruent with that observed in Tead1-icKO mouse hearts — suggesting a similar mechanistic function for Tead1 as a regulator of SERCA activity in both rodent and human CM. We then directly tested if TEAD1 regulates I-1 in human induced-pluripotent stem cell–derived CMs (hIPSC-CMs) (Supplemental Figure 8 and Supplemental Video 3). Pharmacologically blocking TEAD1 activity in differentiated hIPSC-CMs by verteporfin (VP) (35) caused a decrease in SERCA2 and phospho-Ser16-PLN and a complete loss of I-1 protein level (Figure 7D), accompanied by a striking loss of spontaneous contraction (Supplemental Video 4). This strongly suggested that, in hiPSC-CMs, TEAD1 activity — similar to that in mouse CM — is required for SERCA2 and I-1 expression, as well as for the phosphorylation of PLN.
Figure 7. TEAD1 is required for normal SERCA2a, Inhibitor-1, and phospholamban (PLN) phosphorylation in human cardiomyocytes.
(A) Box-whisker plots showing the expression of TEAD1 in human heart samples from idiopathic DCM patients (GSE 1145), presented as mean ± SEM, analyzed by Student’s t test. ***P < 0.001. (B) Enrichment plots showing a strong association between Tead1-icKO gene dataset and the enrichment of published human DCM GSE dataset signature genes. NES, normalized enrichment score. (C) Immunoblot of human heart ventricle samples from normal control individuals and patients with DCM. Associated 3-D scatter plot of the correlation among protein expression of TEAD1, I-1, and pPLNSer16/PLN ratio. Pearson’s correlation was used to determine P values, shown in adjoining table. (D) Representative immunoblot from human induced pluripotent stem cell–derived cardiomyocytes (hIPSC-CMs) treated with verteporfin (VP) 2 μg/ml overnight. The quantification of the protein expression fold changes is plotted from 3 independent experiments. p/tPLN, ratio of phospho to total PLN.
Discussion
In this study, we discovered a role of Tead1 in maintaining adult cardiac contractility, and its loss-of-function leads to severe fatal DCM. We demonstrate that Tead1 is required for normal SR calcium cycling to sustain normal excitation-contraction coupling by directly regulating the transcription and activity of SERCA2a. Mechanistically, Tead1 directly enhances SERCA2a activity by modulating the phosphorylation status of Pln by directly activating the transcription of I-1 and decreasing the activity of SR-associated PP1. The continued expression of Tead1 in postproliferative CMs, thus, appears to be critical and nonredundant.
Tead1 is evolutionarily highly conserved and ubiquitously expressed, with high expression in cardiac muscle. Tead1 has been shown to be important in myogenesis via its binding to malonyl-coA-acyl carrier protein transacylase (MCAT) elements in the promoters of many muscle-specific genes (19, 36, 37). Tead1 and its upstream regulators have been shown to be critical in CM proliferation during embryonic and perinatal periods (8, 22, 23), with recent studies also demonstrating the role of the mammalian Hippo-Tead1 pathway in CM regeneration after injury (38, 39). Indeed, embryonic deletion of Tead1 (16) or a CM-specific deletion of its coactivators Yap and Taz (8), result in embryonic or perinatal lethality, while Yap regulation of CM proliferation was shown to be Tead1 dependent (23). Highlighting the importance of Tead1 in the heart, an earlier study also demonstrated that Tead1, along with Mef2 and Foxo, directly targets myocardin during cardiovascular development (40). Furthermore, other studies identified a cardiac transcription network consisting of Gata4, Mef2a, Nkx2-5, and Srf (20, 40), with an enrichment of Tead1-binding motifs within the binding regions of these transcriptional factors, along with evidence of physical interaction between Tead1 and these factors (20). Similarly, Tead1 was also found to be part of an enhancer cluster composed of Tbx20, Esrr, Creb1, and Mef2a on cardiac-specific genes (21). A recent study demonstrated that deacetylated Vgll4 — a corepressor of Tead1 that disrupts Yap-Tead1 binding, binds, and mediates the degradation of Tead protein — may play a significant role in inhibiting Yap-Tead1–mediated early postnatal CM proliferation (18). All this attests to the critical role of Tead1 in proliferating CMs, but its specific requirement and function in ACMs is unknown. In this study, we demonstrate the nonredundant role of Tead1 in excitation-contraction coupling in ACMs by direct transcriptional activation of SERCA2a and I-1, two critical components of the SR calcium cycling–related genes. In addition, our microarray data also suggest Tead1 has a broader rol×e in transcriptional regulation of other genes important in cardiac function, including mitochondrial- and sarcomere-encoding genes. This is in keeping with Tead1 being a part of a cluster of transcription factor–regulating enhancers in cardiac-specific gene expression (20, 21).
As a coactivator, Yap lacks a DNA-binding domain and is brought to gene promoters/enhancers by partner transcription factors, such as Tead, Runx, Smad, and others, among which the Tead family of transcription factors have been identified as potent binding partners of Yap in CMs. von Gise et al. showed that the interaction with Tead is required for Yap transcription regulation of CM proliferation (23). Lin et al. showed that CM overexpression of YAP-S94A, which lacks the ability to interact with and activate Tead, abolished YAP-mediated mitogenic activity (41). While it is a possibility that, in the absence of Tead1, Yap could potentially redistribute and have other Tead1-independent effects contributing to the phenotype seen in Tead1-icKO hearts, these and other studies strongly indicate that it is unlikely to be a significant contributor to the observed phenotype in Tead1-icKO hearts.
Tsika et al. observed that transgenic overexpression of Tead1, driven by the MCK promoter, in mouse hearts (42) resulted in a significant upregulation of the fetal gene program, including β-MHC. This persistent upregulation of β-MHC, which has a lower power output than the adult α-MHC, was thought to contribute to the decreased cardiac contractility at 10 months of age and to the observed poor tolerance to pressure overload. While, a decrease in Serca2a and Pln phosphorylation was seen in these mice with heart failure, it is unclear if these were secondary to heart failure. Interestingly, both Tead1-overexpressing transgenic (42) and Tead1-deleted mice from our study displayed decreased contractile function. Consistent with the Tsika et al. study that Tead1 overexpression upregulated β-MHC, Tead1-deletion, in our study, led to a significant decrease in β-MHC (data not shown), suggesting that Tead1 is a potent regulator of β-MHC. Whether this regulation of β-MHC by Tead1 is direct will require further experimentation. Our ex vivo and in vivo experiments demonstrate conclusively that Tead1 regulation of SERCA2a and the phosphorylation of Pln is cell autonomous. Taken together, the findings from both of these studies strongly suggest that higher levels of Tead1 protein, such as in the embryonic period or with transgenic overexpression, induce the fetal gene program, while a reduced but persistent Tead1 expression in mature ACMs is required for excitation-contraction coupling and normal contractile function, via coordinated regulation of SERCA2a expression and activity.
Aberrant Ca2+ cycling constitutes an important mechanism of heart failure. Diminished expression of SERCA2a is commonly observed in experimental and human heart failure (43, 44). While recent reports suggest posttranscriptional (45) and posttranslational (46) mechanisms, the underlying mechanisms are still not fully understood. Though Tead1, via its MCAT element binding, has been demonstrated to regulate striated muscle–specific gene expression and differentiation, its role in CM SERCA2a expression has not been explored. In this study, we demonstrate that Tead1 is required for normal SERCA2a expression in both mouse and human CMs, providing a possible mechanism for the downregulation of SERCA2a in human heart failure, as there is also a trend for Tead1 downregulation in human heart failure.
The primary regulator of SERCA2a activity is its inhibitor Pln under basal conditions. Upon β-adrenergic stimulation, Pln is phosphorylated at Ser16 and Thr17 by PKA and CamKII, respectively, leading to a decreased affinity of Pln for SERCA2a, with a consequent increase in Ca2+ affinity of SERCA2a, thus promoting SR Ca2+ uptake (47) and initiating diastolic relaxation. Other studies have also implicated the critical role of PP1 phosphatase activity in regulating the phosphorylation of Pln, with PP1 regulatory subunit I-1 laying a central role. Indeed, decreased I-1 levels with increased PP1 activity and decreased Pln phosphorylation have been reported in experimental and human heart failure (48, 49), though the underlying molecular mechanisms remain unclear, while augmenting I-1 action has had beneficial effects on cardiac contractility in experimental heart failure (50). Here, we show that Tead1 regulates this critical regulator pathway by directly regulating I-1 transcription. This places Tead1 with a dual action of augmenting both SERCA2a expression and its activity, highlighting its nonredundant role in ACM function.
While many of the regulatory mechanisms are conserved in mouse and human heart failure, there still remain important, currently unresolved questions. For instance, SERCA2a overexpression enhances cardiac contractility in vivo (51, 52) in experimental models and in initial human studies (53); the efficacy of this approach could not be confirmed in a larger human heart failure study (54). Although, the regulation of SERCA2a activity by Pln is well established, certain loss-of-function Pln mutations (55), associated with familial DCM, raise the possibility of differential regulatory mechanisms in human CMs. Many reasons have been proposed for these differences (56), including possible fundamental differences in human and rodent CMs, stressing the importance of studying both simultaneously to increase translation of mouse studies to human diseases. In keeping with this, our studies provide preliminary evidence that TEAD1 has a critical role, similar to mouse CM, in regulating SERCA2a, Pln phosphorylation, and I-1 in human hearts and hiPS-derived CMs. Though our study was not designed to answer whether the tendency for decreased TEAD1 protein levels in human heart failure is primary or secondary, the studies in hiPS-derived CM suggest that these changes in TEAD1 would perpetuate the pathophysiology of heart failure by further diminishing SERCA2a expression and activity, raising the possibility that therapeutic strategies to restore TEAD1 levels may be significantly beneficial in slowing the downward spiral — which is often seen in end-stage heart failure — by simultaneously enhancing SERCA2a expression and activity.
This study demonstrates that Tead1 has a cell-autonomous and nonredundant role in ACM excitation-contraction coupling, beyond the currently established Yap-facilitated activation of cellular proliferation and survival. These distinct functions, based on developmental context, are in keeping with similar multitasking seen in other cardiac transcription factors, such as GATA4, NKX2-5, MEF-2A, and SRF (40).
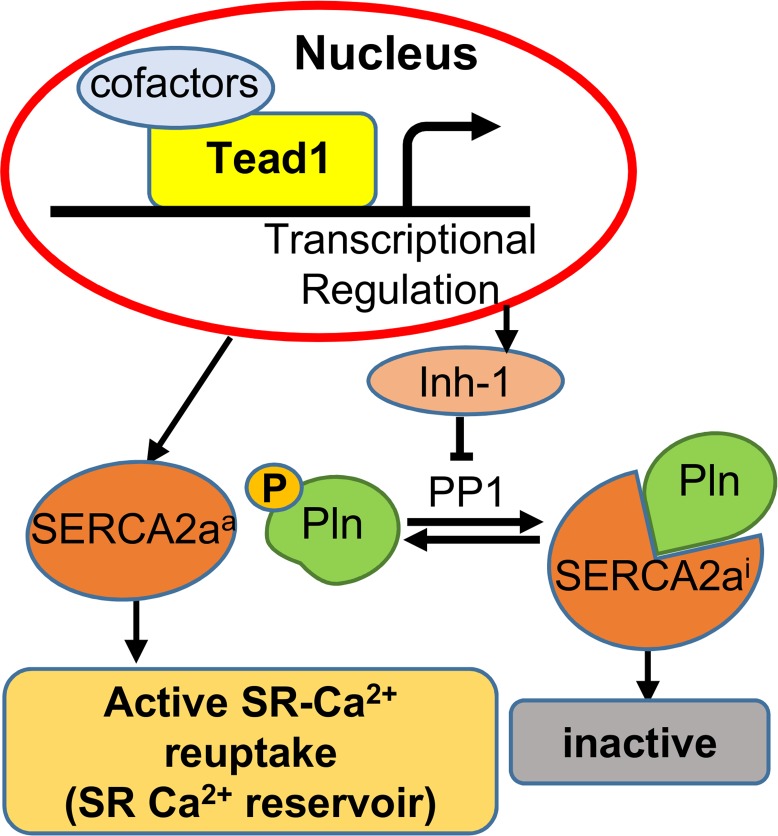
Taken together, we demonstrated that Tead1 is essential in adult cardiac function via maintenance of CM calcium homeostasis, not only by direct transcriptional regulation of SERCA2a expression, but also by enhancing the transcription of I-1 (schematic model shown in Figure 8). We further showed that TEAD1 regulation of PLN, SERCA2a, and I-1 are conserved in human CMs. Given that Serca2a expression and activity are frequently diminished in heart failure, as is I-1 regulation of phospho-PLN (49), our data demonstrating Tead1 as a direct transcriptional activator of SERCA2a and I-1 delineate heretofore unrecognized mechanisms to our knowledge and, thus, could lend themselves for targeted therapy.
Figure 8. Schematic model for Tead1 regulation of adult cardiomyocyte SR-calcium cycling.
Methods
Animal experiments.
The conditional KO of Tead1 was achieved using KO first strategy (reporter-tagged insertion with conditional potential) by Knockout Mouse Project (KOMP; project ID 34183; clone EPD0207_2_H11; see Supplemental Methods). Tead1-icKO was achieved by crossing Tead1F/F mice with Myh6-CreER mice (TM-inducible Cre recombinase under the control of Myh6 CM–specific promoter; the Jackson Laboratory). Homologous recombination in CMs was achieved by administering 3 weeks of TM diet (TD.140450, Harlan Lab) at the dose of 125 mg TM kg–1 diet to Tead1F/FMyh6-CreER mice. Mice were maintained under standard 12-hour light-dark cycles with ad libitum access to food and water, unless specified otherwise.
Mice echocardiography.
Under 1.5% isoflurane anesthesia, echocardiographic examination was conducted on the mice using VevoR 2100 System (VisualSonics) with body temperature and heart rate monitored. End systolic and diastolic dimensions were obtained from M-mode ultrasound; EF and FS were calculated accordingly. Echocardiograms were performed blinded to genotype and treatment group.
Isolation, culture, and ex vivo KO in mouse NCMs.
NCMs were isolated from the hearts of 0- to 3-day-old neonatal mice as described previously (57). Briefly, ventricles from neonates were dissected, followed by 1.5 mg/ml Collagenase/Dispase (Roche Diagnostics) digestion. The isolated cells were plated in plating medium that contains 65% high glucose DMEM (GenDEPOT), 19% M199 (HyClone), 10% horse serum (HS), 5% FBS, and antibiotic-antimycotic (Gibco) at 37°C and 5% CO2 overnight. Then, cells were changed to maintaining medium that contains 78% high glucose DMEM, 19% M199, 4% HS, and antibiotic-antimycotic. To achieve ex vivo Tead1 KO, NCMs isolated from Tead1F/F neonates were transduced with the indicated adenoviruses or lentivirus at 50 MOI. Adenoviruses encoding Cre recombinase (Cre) or β-galactosidase (lacZ) were generated by Gene Vector Core in the Baylor College of Medicine Advanced Technology Core Laboratories.
Isolation and confocal Calcium imaging of mouse ACMs.
ACMs were isolated from anesthetized mice as described previously (58). Briefly, mouse hearts were cannulated via ascending aorta and then perfused on a Langendorff’s apparatus with 0 Ca2+ Tyrode solution (137 mmol/l NaCl, 5.4 mmol/l KCl, 1 mmol/l MgCl2, 5 mmol/l HEPES, 10 mmol/l glucose, 3 mmol/l NaOH, pH 7.4; all chemicals are from Sigma-Aldrich, unless indicated otherwise), followed by 20 μg/ml Liberase (Roche) digestion, and then dissociated into single cells. ACMs were equilibrated in Kraftbruhe (KB) solution (90 mmol/l KCl, 30 mmol/l K2HPO4, 5 mmol/l MgSO4, 5 mmol/l pyruvic acid, 5 mmol/l β-hydroxybutyric acid, 5 mmol/l creatine, 20 mmol/l taurine, 10 mmol/l glucose, 0.5 mmol/l EGTA, 5 mmol/l HEPES, pH 7.2). For confocal imaging, ACMs were loaded with 2 μmol/l Fluo-4/AM (Invitrogen) in 1.8 mmol/l Ca2+ Tyrode solution in for 30 minutes at room temperature; then, cells were loaded on a laser scanning confocal microscope (LSM510, Carl Zeiss) with a ×40 oil immersion objective. Fluorescence images were recorded in line-scan mode with 1,024 pixels per line at 500 Hz. After being paced at 1 Hz for 20 seconds, rod-shaped myocytes showing clear striation and normal contractility were selected for further experiments. Steady state SR Ca2+ content was calculated via transient amplitude (F1/F0) induced by rapid application of caffeine (10 mmol/l) after pacing.
SERCA activity and myocardial PKA activity assays.
Cardiac SR vesicles were prepared as previously described (59). The activity of SR-associated PP1 was measured using a fluorometric assay based on the ProFluorR Ser/Thr Phosphatase (PPase) Assay kit (V1260, Promega) following the manufacturer’s protocol. Briefly, cardiac SR was IP with protein G-agarose beads by PP1c antibody (sc-2003, Santa Cruz Biotechnology Inc.). The IP product was diluted in PPase dilution solution and incubated with the peptide solution, followed by incubation with the protease solution. Stabilizer solution was then added, and the PP1 activity was determined with FLUOstar Omega plate reader (BMG Labtech) by measuring the fluorescence of the R110 substrate at 485/530 nm. The fluorescence change correlates with PP1 activity, and this was standardized to the quantity of PP1c protein used for the assay.
SR-associated SERCA hydrolytic activity was determined by a colorimetric method (60, 61). The reaction was carried in the assay buffer contained 40 mmol/l Tris, 80 mmol/l NaCl, 8 mmol/l MgAc2, 1 mmol/l EDTA, pH7.5. SR protein (1 μg) was incubated with 200 μmol/l CaCl2 and 3 μmol/l A23187 ionophore for 10 minutes, and the reaction was initiated by addition of 1 mmol/l ATP and incubated at 37°C for 30 minutes. The production of phosphate that is proportional to SERCA activity was measured using ATPase activity assay kit (MAK113, Sigma-Aldrich) following manufacturer’s protocol. Briefly, the malachite green reagent provided in the kit formed a dark green color with free phosphate, which was colorimetrically measured at 620 nm.
The PKA activity was measured using a nonradioactive assay kit (ADI-EKS-390A, Enzo Life Sciences) according to the manufacturer’s protocol. Briefly, mouse ventricle protein lysate was incubated with ATP in the PKA substrate microtiter plate at 30°C for 1 hour, followed by incubation with phosphospecific substrate antibody (primary) and anti–rabbit HRP conjugated IgG (secondary). The assay was developed with tetramethylbenzidine (TMB) substrate and stopped with acid stop solution, the activity was determined with a microplate reader by measuring the absorbance at 450 nm. Purified active PKA was served as positive control. The relative PKA activity was calculated using the following formula: (average absorbance[sample] – average absorbance[blank])/quantity of protein per assay.
Generation of hiPSCs (M22 hiPSC line) and directed differentiation of CMs.
To initiate reprogramming, primary adult human male dermal fibroblasts (Zen-Bio) were infected with nonintegrating Sendai viruses expressing OCT4, SOX2, KLF4, and c-MYC using manufacturer’s guidelines (CytoTune-iPS 2.0, Thermo Fisher). Clonal hiPSC colonies were manually picked and expanded under feeder-free conditions using hESC-qualified Matrigel (Corning) and TeSR-E8 medium (Stemcell Technologies). Clone 5 was shown to have a normal male karyotype at passage 5 and subsequently used for all differentiation experiments. Clone 5 hiPSCs were differentiated into embryoid bodies for 3 weeks, and they were shown to express tri-lineage markers using the TaqMan hPSC Scorecard assay (Applied Biosystems). This characterization was performed by the Human Stem Cell Core at Baylor College of Medicine.
M22 hiPSCs were differentiated into CMs using the Activin A-BMP4 protocol as previously described (Supplemental Figure 8A; ref. 62). Small beating areas were seen starting on day 7 and more synchronized rhythmic beating in larger areas was observed throughout the culture. Cells were dissociated into single cells after 4 weeks of differentiation and allowed to attach in Matrigel-coated cover slips for immunostaining of CM markers.
Patients and tissue collection.
Heart-failure tissues were collected from 4 patients (Supplemental Table 3) undergoing left ventricular assist device (LVAD) insertion or heart transplantation at the Houston Methodist Hospital (Houston, Texas, USA). Tissues collected in the operating rooms were flash frozen in liquid nitrogen and preserved for further analysis.
Statistics.
The data are presented as means ± SEM. Significance tests were determined by the 2-tailed, unpaired Student’s t test or 1-way ANOVA with Tukey’s post hoc test. P < 0.05 was considered statistically significant. GraphPad Prism 5.0 software was used for statistical analyses.
Study approval.
All animal procedures were approved by the IACUC of Baylor College of Medicine conforming to the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011). The investigation conformed with the principles outlined in the Declaration of Helsinki and was approved by the Houston Methodist Hospital’s IRB (IRB[2]0511–0100). Signed consent was obtained from each patient before enrollment for the study.
Author contributions
RL designed and conducted the experiments, performed data analyses, interpreted data, and wrote the manuscript. JL, BSK, QW, SKB, NB, JJK, JD, AZ, and SL helped with specific experiments and data analysis. AAG, DJH, JFM, GGR, JAT, CC, and XW provided critical scientific input, data analysis, and logistical support and edited the manuscript. VKY and MM supervised the project, designed experiments, interpreted data, and edited the manuscript.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH K08-HL091176 (to MM); Veterans Affairs Office of Research and Development–Biomedical Laboratory Research and Development-I01BX002678, and NIH R01 DK097160 (to VKY); and NIH R01-HL089598, R01-HL091947, R01-HL117641, and R41-HL129570 (to XW). This work was also supported by the Muscular Dystrophy Association (to XW) and the American Heart Association 13EIA14560061 (to XW), and NIH R01-AR061370 (to GGR), as well as by the BCM Advanced Core Labs (NIH U54-HG006348, P30-DK079638, P30-CA125123, P30-DK076938). RL is a National Heart, Lung, and Blood Institute Scholarship recipient.
Version 1. 08/23/2017
Electronic publication
Footnotes
M. Moulik’s present address is: Department of Pediatrics/Division of Cardiology, University of Pittsburgh, Pittsburgh, USA.
V.K. Yechoor’s present address is: Department of Medicine, Division of Endocrinology and Metabolism, University of Pittsburgh, Pittsburgh, USA.
Conflict of interest statement: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(17):e93343. https://doi.org/10.1172/jci.insight.93343.
Contributor Information
Ruya Liu, Email: liuruya212@gmail.com.
Jeongkyung Lee, Email: jkleebio@gmail.com.
Qiongling Wang, Email: qiongliw@bcm.edu.
Nikhil Balasubramanyam, Email: balasubramanyam.nikhil@gmail.com.
Jianrong Dong, Email: Jianrong.Dong@bcm.edu.
Aijun Zhang, Email: AZHANG@houstonmethodist.org.
Shumin Li, Email: sli2@houstonmethodist.org.
James F. Martin, Email: jfmartin@bcm.edu.
George G. Rodney, Email: rodney@bcm.edu.
Cristian Coarfa, Email: coarfa@bcm.edu.
Xander H.T. Wehrens, Email: wehrens@bcm.edu.
Mousumi Moulik, Email: moulikm@pitt.edu.
References
- 1.Liu L, Eisen HJ. Epidemiology of heart failure and scope of the problem. Cardiol Clin. 2014;32(1):1–8. doi: 10.1016/j.ccl.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Towbin JA, et al. Incidence, causes, and outcomes of dilated cardiomyopathy in children. JAMA. 2006;296(15):1867–1876. doi: 10.1001/jama.296.15.1867. [DOI] [PubMed] [Google Scholar]
- 3.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10(9):531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 4.Bozkurt B, et al. Current diagnostic and treatment strategies for specific dilated cardiomyopathies: a scientific statement from the American Heart Association. Circulation. 2016;134(23):e579–e646. doi: 10.1161/CIR.0000000000000455. [DOI] [PubMed] [Google Scholar]
- 5.Watkins H, Ashrafian H, Redwood C. Inherited cardiomyopathies. N Engl J Med. 2011;364(17):1643–1656. doi: 10.1056/NEJMra0902923. [DOI] [PubMed] [Google Scholar]
- 6.Yu FX, Zhao B, Guan KL. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163(4):811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plouffe SW, Hong AW, Guan KL. Disease implications of the Hippo/YAP pathway. Trends Mol Med. 2015;21(4):212–222. doi: 10.1016/j.molmed.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xin M, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A. 2013;110(34):13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin Z, et al. Pi3kcb links Hippo-YAP and PI3K-AKT signaling pathways to promote cardiomyocyte proliferation and survival. Circ Res. 2015;116(1):35–45. doi: 10.1161/CIRCRESAHA.115.304457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heallen T, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332(6028):458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Q, Li L, Zhao B, Guan KL. The hippo pathway in heart development, regeneration, and diseases. Circ Res. 2015;116(8):1431–1447. doi: 10.1161/CIRCRESAHA.116.303311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeda S, Sadoshima J. Regulation of myocardial cell growth and death by the Hippo pathway. Circ J. 2016;80(7):1511–1519. doi: 10.1253/circj.CJ-16-0476. [DOI] [PubMed] [Google Scholar]
- 13.Jiang SW, Desai D, Khan S, Eberhardt NL. Cooperative binding of TEF-1 to repeated GGAATG-related consensus elements with restricted spatial separation and orientation. DNA Cell Biol. 2000;19(8):507–514. doi: 10.1089/10445490050128430. [DOI] [PubMed] [Google Scholar]
- 14.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30(1):1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Yu A, Yu FX. The Hippo pathway in tissue homeostasis and regeneration. Protein Cell. 2017;8(5):349–359. doi: 10.1007/s13238-017-0371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Friedrich GA, Soriano P. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 1994;8(19):2293–2301. doi: 10.1101/gad.8.19.2293. [DOI] [PubMed] [Google Scholar]
- 17.Chen HH, Mullett SJ, Stewart AF. Vgl-4, a novel member of the vestigial-like family of transcription cofactors, regulates alpha1-adrenergic activation of gene expression in cardiac myocytes. J Biol Chem. 2004;279(29):30800–30806. doi: 10.1074/jbc.M400154200. [DOI] [PubMed] [Google Scholar]
- 18.Lin Z, et al. Acetylation of VGLL4 regulates Hippo-YAP signaling and postnatal cardiac growth. Dev Cell. 2016;39(4):466–479. doi: 10.1016/j.devcel.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida T. MCAT elements and the TEF-1 family of transcription factors in muscle development and disease. Arterioscler Thromb Vasc Biol. 2008;28(1):8–17. doi: 10.1161/ATVBAHA.107.155788. [DOI] [PubMed] [Google Scholar]
- 20.He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci U S A. 2011;108(14):5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen T, et al. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J Clin Invest. 2011;121(12):4640–4654. doi: 10.1172/JCI59472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135(24):4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 23.von Gise A, et al. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc Natl Acad Sci U S A. 2012;109(7):2394–2399. doi: 10.1073/pnas.1116136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahuja P, Sdek P, MacLellan WR. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev. 2007;87(2):521–544. doi: 10.1152/physrev.00032.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sohal DS, et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89(1):20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 26.Koitabashi N, et al. Avoidance of transient cardiomyopathy in cardiomyocyte-targeted tamoxifen-induced MerCreMer gene deletion models. Circ Res. 2009;105(1):12–15. doi: 10.1161/CIRCRESAHA.109.198416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lexow J, Poggioli T, Sarathchandra P, Santini MP, Rosenthal N. Cardiac fibrosis in mice expressing an inducible myocardial-specific Cre driver. Dis Model Mech. 2013;6(6):1470–1476. doi: 10.1242/dmm.010470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersson KB, Winer LH, Mørk HK, Molkentin JD, Jaisser F. Tamoxifen administration routes and dosage for inducible Cre-mediated gene disruption in mouse hearts. Transgenic Res. 2010;19(4):715–725. doi: 10.1007/s11248-009-9342-4. [DOI] [PubMed] [Google Scholar]
- 29.Frank KF, Bölck B, Erdmann E, Schwinger RH. Sarcoplasmic reticulum Ca2+-ATPase modulates cardiac contraction and relaxation. Cardiovasc Res. 2003;57(1):20–27. doi: 10.1016/S0008-6363(02)00694-6. [DOI] [PubMed] [Google Scholar]
- 30.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4(7):517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 31.Gustavsson M, et al. Allosteric regulation of SERCA by phosphorylation-mediated conformational shift of phospholamban. Proc Natl Acad Sci U S A. 2013;110(43):17338–17343. doi: 10.1073/pnas.1303006110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacLennan DH, Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol. 2003;4(7):566–577. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 33.MacDougall LK, Jones LR, Cohen P. Identification of the major protein phosphatases in mammalian cardiac muscle which dephosphorylate phospholamban. Eur J Biochem. 1991;196(3):725–734. doi: 10.1111/j.1432-1033.1991.tb15871.x. [DOI] [PubMed] [Google Scholar]
- 34.Wittköpper K, Dobrev D, Eschenhagen T, El-Armouche A. Phosphatase-1 inhibitor-1 in physiological and pathological β-adrenoceptor signalling. Cardiovasc Res. 2011;91(3):392–401. doi: 10.1093/cvr/cvr058. [DOI] [PubMed] [Google Scholar]
- 35.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26(12):1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta MP, Amin CS, Gupta M, Hay N, Zak R. Transcription enhancer factor 1 interacts with a basic helix-loop-helix zipper protein, Max, for positive regulation of cardiac alpha-myosin heavy-chain gene expression. Mol Cell Biol. 1997;17(7):3924–3936. doi: 10.1128/MCB.17.7.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joshi S, et al. TEAD transcription factors are required for normal primary myoblast differentiation in vitro and muscle regeneration in vivo. PLoS Genet. 2017;13(2):e1006600. doi: 10.1371/journal.pgen.1006600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heallen T, et al. Hippo signaling impedes adult heart regeneration. Development. 2013;140(23):4683–4690. doi: 10.1242/dev.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morikawa Y, et al. Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice. Sci Signal. 2015;8(375):ra41. doi: 10.1126/scisignal.2005781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlesinger J, et al. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genet. 2011;7(2):e1001313. doi: 10.1371/journal.pgen.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Z, et al. Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ Res. 2014;115(3):354–363. doi: 10.1161/CIRCRESAHA.115.303632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsika RW, et al. TEAD-1 overexpression in the mouse heart promotes an age-dependent heart dysfunction. J Biol Chem. 2010;285(18):13721–13735. doi: 10.1074/jbc.M109.063057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de la Bastie D, et al. Function of the sarcoplasmic reticulum and expression of its Ca2(+)-ATPase gene in pressure overload-induced cardiac hypertrophy in the rat. Circ Res. 1990;66(2):554–564. doi: 10.1161/01.RES.66.2.554. [DOI] [PubMed] [Google Scholar]
- 44.Mercadier JJ, et al. Altered sarcoplasmic reticulum Ca2(+)-ATPase gene expression in the human ventricle during end-stage heart failure. J Clin Invest. 1990;85(1):305–309. doi: 10.1172/JCI114429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahlquist C, et al. Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature. 2014;508(7497):531–535. doi: 10.1038/nature13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kho C, et al. SUMO1-dependent modulation of SERCA2a in heart failure. Nature. 2011;477(7366):601–605. doi: 10.1038/nature10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simmerman HK, Jones LR. Phospholamban: protein structure, mechanism of action, and role in cardiac function. Physiol Rev. 1998;78(4):921–947. doi: 10.1152/physrev.1998.78.4.921. [DOI] [PubMed] [Google Scholar]
- 48.Neumann J, et al. Increased expression of cardiac phosphatases in patients with end-stage heart failure. J Mol Cell Cardiol. 1997;29(1):265–272. doi: 10.1006/jmcc.1996.0271. [DOI] [PubMed] [Google Scholar]
- 49.El-Armouche A, Pamminger T, Ditz D, Zolk O, Eschenhagen T. Decreased protein and phosphorylation level of the protein phosphatase inhibitor-1 in failing human hearts. Cardiovasc Res. 2004;61(1):87–93. doi: 10.1016/j.cardiores.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 50.Nicolaou P, Hajjar RJ, Kranias EG. Role of protein phosphatase-1 inhibitor-1 in cardiac physiology and pathophysiology. J Mol Cell Cardiol. 2009;47(3):365–371. doi: 10.1016/j.yjmcc.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.del Monte F, et al. Improvement in survival and cardiac metabolism after gene transfer of sarcoplasmic reticulum Ca(2+)-ATPase in a rat model of heart failure. Circulation. 2001;104(12):1424–1429. doi: 10.1161/hc3601.095574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lyon AR, et al. SERCA2a gene transfer decreases sarcoplasmic reticulum calcium leak and reduces ventricular arrhythmias in a model of chronic heart failure. Circ Arrhythm Electrophysiol. 2011;4(3):362–372. doi: 10.1161/CIRCEP.110.961615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaski BE, et al. Calcium upregulation by percutaneous administration of gene therapy in cardiac disease (CUPID Trial), a first-in-human phase 1/2 clinical trial. J Card Fail. 2009;15(3):171–181. doi: 10.1016/j.cardfail.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greenberg B, et al. Calcium upregulation by percutaneous administration of gene therapy in patients with cardiac disease (CUPID 2): a randomised, multinational, double-blind, placebo-controlled, phase 2b trial. Lancet. 2016;387(10024):1178–1186. doi: 10.1016/S0140-6736(16)00082-9. [DOI] [PubMed] [Google Scholar]
- 55.Haghighi K, et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc Natl Acad Sci U S A. 2006;103(5):1388–1393. doi: 10.1073/pnas.0510519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kranias EG, Hajjar RJ. The Phospholamban Journey 4 decades after setting out for Ithaka. Circ Res. 2017;120(5):781–783. doi: 10.1161/CIRCRESAHA.116.310007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ehler E, Moore-Morris T, Lange S. Isolation and culture of neonatal mouse cardiomyocytes. J Vis Exp. 2013;79 doi: 10.3791/50154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Q, Wang W, Wang G, Rodney GG, Wehrens XH. Crosstalk between RyR2 oxidation and phosphorylation contributes to cardiac dysfunction in mice with Duchenne muscular dystrophy. J Mol Cell Cardiol. 2015;89(pt B):177–184. doi: 10.1016/j.yjmcc.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wehrens XH, et al. FKBP12. Cell. 2003;113(7):829–840. doi: 10.1016/S0092-8674(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 60.Bartolommei G, Tadini-Buoninsegni F, Hua S, Moncelli MR, Inesi G, Guidelli R. Clotrimazole inhibits the Ca2+-ATPase (SERCA) by interfering with Ca2+ binding and favoring the E2 conformation. J Biol Chem. 2006;281(14):9547–9551. doi: 10.1074/jbc.M510550200. [DOI] [PubMed] [Google Scholar]
- 61.Chu A, Dixon MC, Saito A, Seiler S, Fleischer S. Isolation of sarcoplasmic reticulum fractions referable to longitudinal tubules and junctional terminal cisternae from rabbit skeletal muscle. Meth Enzymol. 1988;157:36–46. doi: 10.1016/0076-6879(88)57066-0. [DOI] [PubMed] [Google Scholar]
- 62.Lian X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 2013;8(1):162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.