Figure 3. Conformational rearrangements of Cdc37 during the Hsp90 cycle.
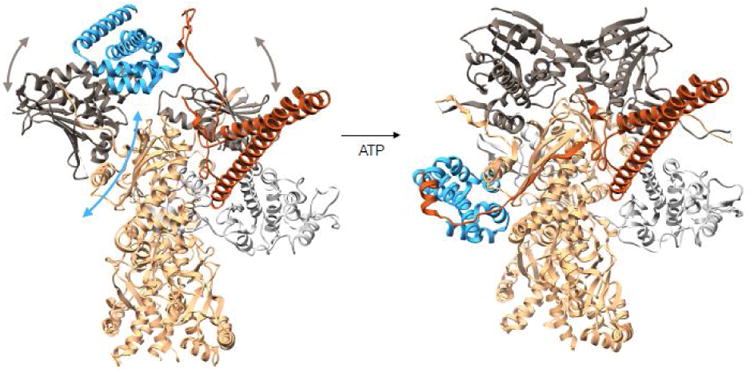
On the left is a modeled complex between Hsp90-Cdc37-Cdk4 where Cdc37-Hsp90 interactions are preserved as in the crystal structure of the fragments (PDB:1US7). The Cdc37 middle domain is colored in blue with the N terminal domain in piper, Hsp90 is in tan with N terminal domains in charcoal and Cdk4 kinase is in light gray. In this state Hsp90's N terminal domains are parted, and the domain of the monomer on the left had to undergo a rotation as to avoid clash between Hsp90's middle domain and Cdc37's middle domain. At this state, we imagine the kinase lobes to be partially open. Upon ATP binding to the Hsp90 N terminal domains, these undergo closure displacing Cdc37 down to the middle domain and stabilizing the kinase in the unfolded state, resulting in the observed cryoEM structure (domain motions are depicted with arrows). Structure on the left corresponds to state (b) in Figure 4 and structure on the right corresponds to state (c) in Figure 4.
