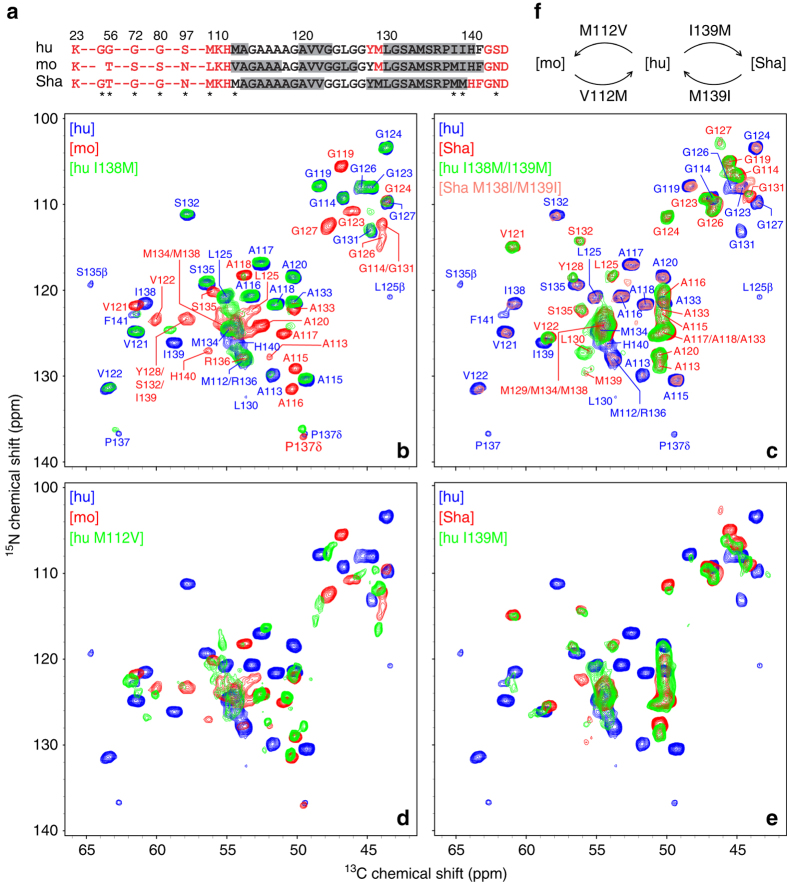
Fig. 1.
Two-dimensional 15N–13Cα solid-state NMR spectra of PrP23-144 amyloid fibrils. a Amino acid sequences of hu, mo, and ShaPrP23-144. Immobile residues, located within the amyloid β-core and observable in solid-state NMR spectra, are shown in black font, and undetectable, conformationally flexible residues are shown in red font. Residues having the highest β-strand propensity according to TALOS-N41 chemical-shift based secondary structure analysis described previously36 are indicated by gray rectangles. The asterisks indicate all residues that are not conserved between the hu, mo, and ShaPrP23-144 sequences. b–e Two-dimensional 15N–13Cα fingerprint NMR spectra of PrP23-144 amyloid fibrils recorded at 500 or 800 MHz 1H frequency, 11.111 kHz MAS rate, and 5 °C. Spectra are shown for a total of eight PrP23-144 amyloids, including the following proteins and their single or double mutants: [hu], [mo], [Sha], [hu M112V], [hu I138M], [hu I139M], [hu I138M/I139M], and [Sha M138I/M139I], as indicated in the insets and by contours of corresponding color. Also shown in corresponding color in b and c are the resonance assignments for [hu], [mo], and [Sha] fibrils32, 36. f Summary of the key amino acid substitutions that, based on the solid-state NMR data in this study, are primarily responsible for the PrP23-144 amyloid β-core adopting [hu], [mo], or [Sha]-like structures