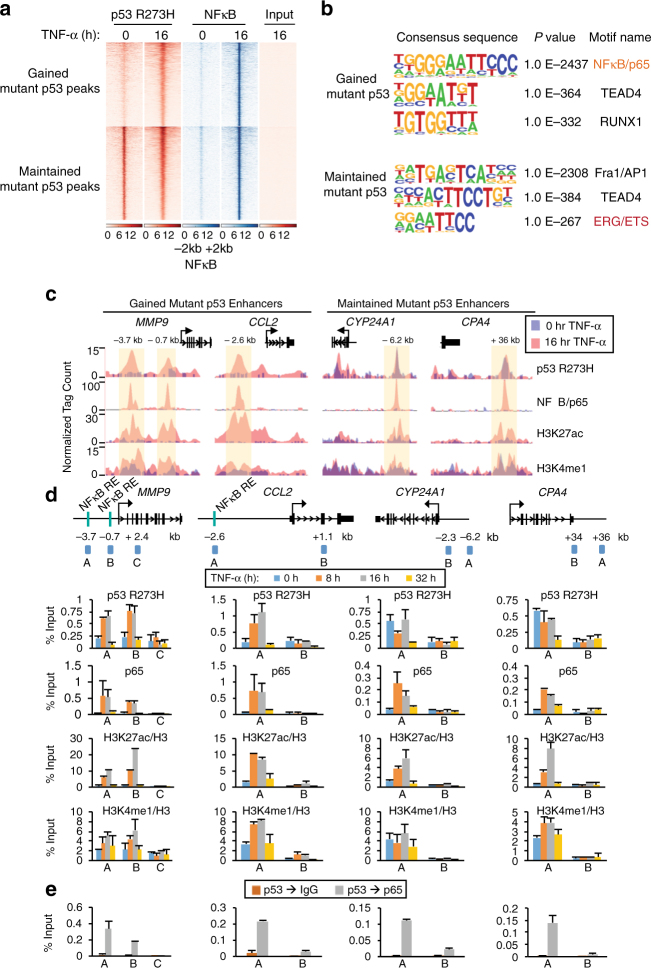
Fig. 2.
Chronic TNF-α signaling alters mutp53 and NFκB binding in colon cancer cells. a Heat maps of p53 R273H and NFκB/p65 ChIP-seq reads in SW480 cells treated with TNF-α for 0 or 16 h. Each row shows ±2 kb centered on p65 peaks, rank-ordered by the intensity of mutp53 and NFκB/p65 peaks and grouped by gained versus maintained mutp53 peaks. b De novo motif analyses of the TNF-gained and maintained mutp53 overlapping NFκB/p65 binding sites as noted in (a). c UCSC genome browser tracks of ChIP-seq signals for p53 R273H, NFκB/p65, H3K27ac, and H3K4me1 at the MMP9, CCL2, CYP24A1, and CPA4 gene loci in untreated (purple) or TNF-α 16 h (pink)-treated SW480 cells. The y axis depicts the ChIP-seq signal and the x axis locates the genomic position with the enhancer regions highlighted in yellow. d Schematics of ChIP-qPCR amplicons and ChIP analyses with the indicated antibodies at the enhancers and non-specific regions of MMP9, CCL2, CYP24A1, and CPA4 gene loci. ChIP-qPCR amplicons were designed to amplify the enhancer (A and B at MMP9 and A at CCL2, CYP24A1, and CPA4) or non-specific (C at MMP9 and B at CCL2, CYP24A1, and CPA4) regions of the target gene loci. ChIP experiments were performed using SW480 cells treated with TNF-α for 0, 8, 16, and 32 h. ChIPs for histone marks were normalized to H3. An average of two independent ChIP experiments that are representative of at least three is shown with error bars denoting the standard error. e Sequential ChIP (re-ChIP) with p53 antibody followed by IgG (control) and NFκB/p65 antibody performed in SW480 cells treated with TNF-α for 16 h. The ChIP-qPCR amplicons are identical to those used in (d). An average of two independent re-ChIP experiments that are representative of at least three is shown with error bars denoting the standard error