Figure 1.
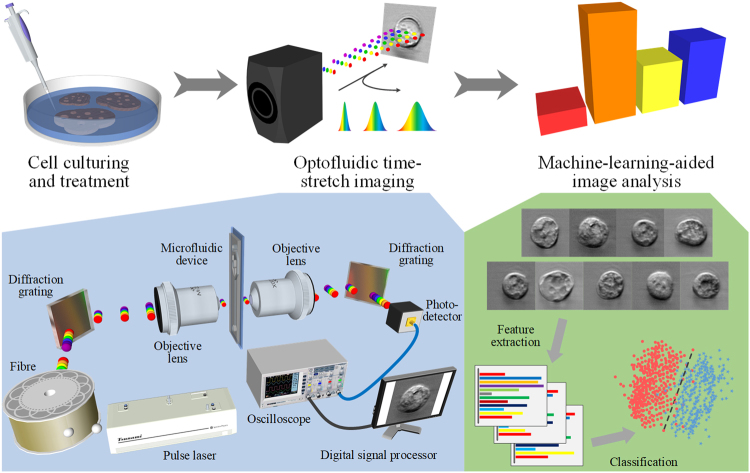
Workflow of the label-free detection of drug-induced morphological variations in cancer cells with optofluidic time-stretch microscopy. The insets show the schematics of optofluidic time-stretch microscopy and machine-learning-aided image analysis. As schematically shown in the figure, the procedure of our method can be divided into three parts: (i) cell culturing and treatment, (ii) optofluidic time-stretch imaging, and (iii) machine-learning-aided image analysis. In the first part, cells of interest are cultured and treated by a drug. In the second part, the treated cells are subject to high-throughput bright-field imaging. In the last part, machine learning algorithm is applied to the images for the identification of their morphological variations induced by drug treatment. While the morphological change in a single cell is miniscule, the large number of bright-field single-cell images can render the morphological change discovered by machine learning statistically significant and robust.