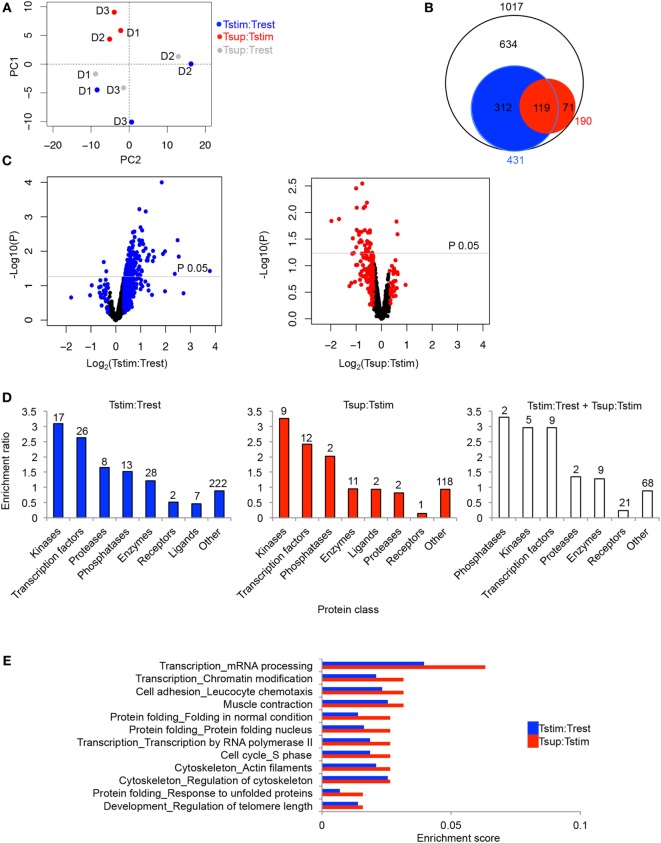
Figure 2.
Activation- and Treg-induced changes in the phosphoproteome of Tcons. Unique phosphopeptides detected in ≥2 out of 3 donors were considered for summary analyses. The ratios reflecting changes upon TCR activation (Tstim:Trest), Treg-mediated suppression (Tsup:Tstim), and suppression compared to the unstimulated state (Tsup:Trest) are represented in blue, red, and gray, respectively. In addition, filtering was done in (B–E) to focus on the regulated phosphopeptides changing ≥25% in respective comparisons. (A) Principle component analysis (PCA) was performed based on the given phosphopeptide ratios for all three donors (D1, D2, D3). (B) Circles represent the total number (white), TCR stimulation-regulated (blue), and suppression-regulated (red) phosphopeptides along with the overlaps. Circle sizes are proportional to the numbers. (C) Volcano plots show stimulation-induced (left) and suppression-induced changes (right). The black dots represent unique phosphopeptides with <25% change. P values were calculated with Bayes moderated t test in limma package in R. log2 of average ratios of phosphopeptides are plotted against −log10 of P values in the particular comparison. (D) Enrichment ratio for respective classes of phosphoproteins regulated upon activation (blue), suppression (red), and jointly upon activation and suppression (white). Enrichment ratio was calculated as ratio between actual number of proteins (numbers above bars) and expected number of proteins enriched in individual categories according to “Enrichment by protein function” analysis in MetaCore. (E) Enrichment scores for process networks enriched (P < 0.05) upon Treg-mediated suppression are paired with corresponding process networks upon TCR stimulation and are arranged according to descending enrichment values in Tsup:Tstim. Enrichment score was calculated as ratio between number of items from the respective MetaCore process present in the lists and total items enlisted under the process. Total number of proteins detected in ≥2 out of 3 donors was set as background.