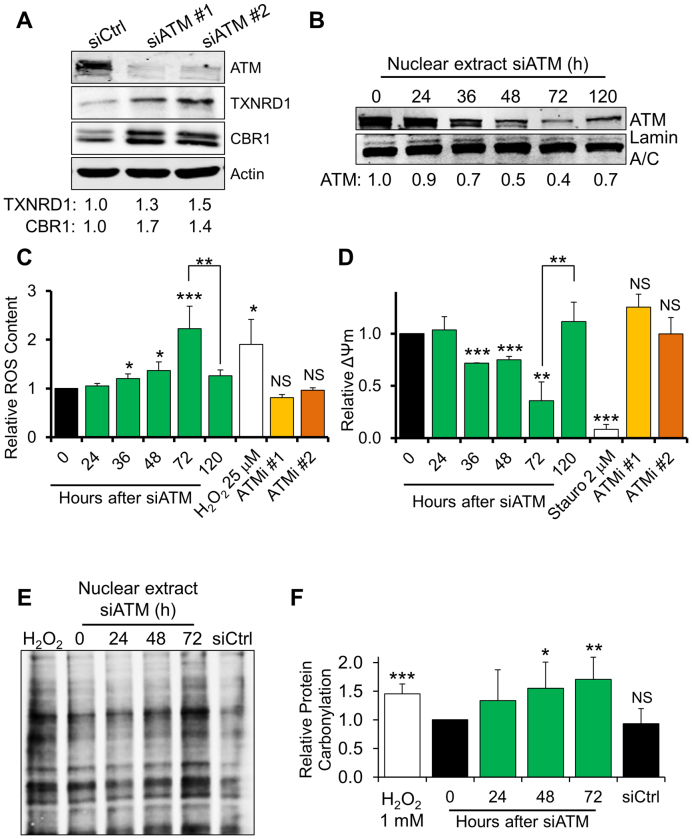
Figure 1.
ATM-depleted fibroblasts progressively accumulate ROS and oxidative protein damage. (A) Representative Western blot analysis on ATM-depleted fibroblasts showing upregulation of proteins involved in the antioxidant response. TIG1 cells were transfected with either a control siRNA (siCtrl) or an ATM-targeting siRNA (siATM#1, siATM #2). Actin was used as loading control. Densitometric quantification of the indicated proteins is reported at the bottom of the gel (N = 2). (B) Western blot analysis on a representative time course depletion of ATM. TIG1 fibroblasts were treated as indicated and ATM expression was monitored in nuclear cell extracts. Lamin A/C was used as loading control. (C) Quantification of ROS content using flow-cytometry in siATM-treated fibroblasts. TIG1 fibroblasts were treated as indicated. H2O2 (25 μM, 30 min) was used as a positive control for induction of ROS. ATM inhibitors (Ku-55933 – ATMi #1 and Ku-60019 – ATMi #2, both 10 μM) were provided fresh every 24 h for a total of 72 h (N = 3). (D) Quantification of mitochondrial membrane potential (ΔΨm) in siATM-treated fibroblasts using flow-cytometry. Cells were treated as in panel C. Staurosporine (2 μM, 2 h) was used as a positive control for mitochondrial membrane depolarisation (N = 3). (E) Detection of protein carbonylation in nuclear extracts obtained from fibroblasts depleted of ATM for the indicated time. Carbonylated proteins were detected by derivatisation with dinitrophenylhydrazone (DNP) followed by Western blot using an anti-DNP antibody. Equal amounts of cell extract were loaded in each lane. H2O2 (1 mM, 30 minutes) was used as a positive control for induction of protein carbonylation. (F) Quantification of protein carbonylation in ATM-depleted fibroblasts; the histogram displays densitometric data from the analysis reported in panel E (N = 6). Results are expressed as mean ± SD from the indicated number (N) of independent experiments: *P < 0.05; **P < 0.01; ***P < 0.001; NS: not significant.