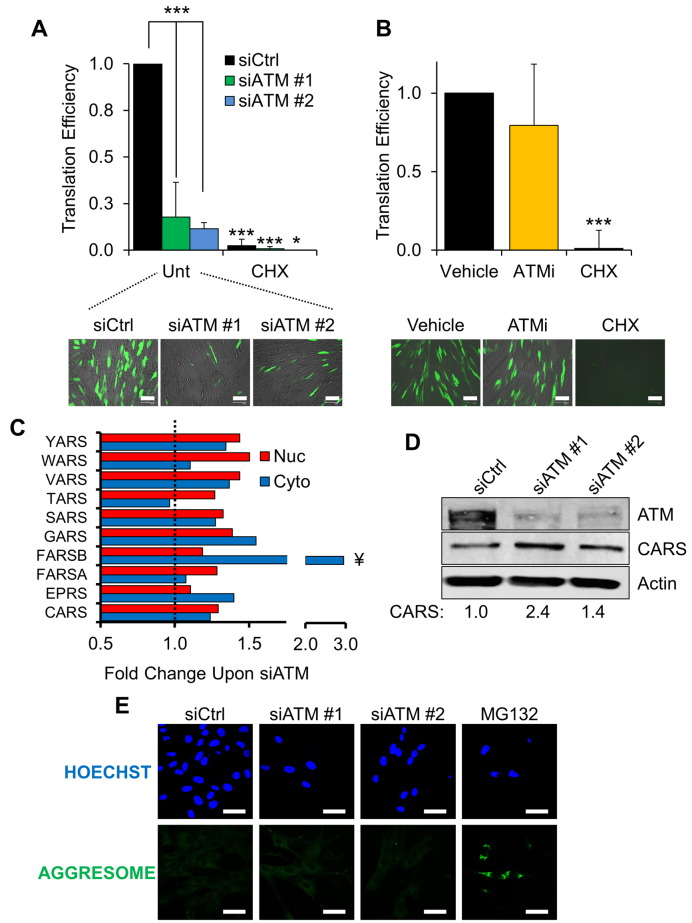
Figure 2.
Fibroblasts lacking ATM show changes in protein biosynthesis. (A) Protein translation assay. TIG1 fibroblasts were transfected with the indicated siRNA and subsequently with an EGFP reporter, as indicated in ‘Materials and Methods’. Translation efficiency was determined by calculating the ratio between the fluorescence of the reporter (bottom panel) and EGFP transcription. ATM-depleted cells show significantly lower translation efficiency than control fibroblasts. Cycloheximide-treated cells (CHX, 25 μg/ml for 24 hours) were used as positive control for translation suppression. Scale bar in the representative micrographs is 100 μm (N = 3). (B) Translation assay carried out as in panel A. Cells were treated with either vehicle (DMSO) or ATM kinase inhibitor (KU-55933, 10 μM for 72 h, fed fresh every 24 h). Scale bar 100 μm (N = 3). (C) Proteomics data showing the fold change in aaRSs expression upon ATM depletion. Red and blue bars show the change in protein expression in the nuclear and cytoplasmic fractions, respectively. Data are expressed as the average fold change calculated between two technical replicates. The dashed line represents the normalised expression level in cells transfected with the control siRNA. ¥: statistically significant hits (Z-score > 2). (D) Validation of the SILAC data using Western blot. The expression level of CARS was measured 72 h after transfection with different ATM-targeting siRNAs. Densitometric quantification of CARS is reported at the bottom (N = 2). (E) Immunofluorescence analysis on TIG1 fibroblasts treated with the indicated siRNA. Aggresome formation was detected as described in ‘Materials and Methods’. MG132 (5 μM for 16 h) was used as positive control for induction of protein aggregates. Nuclei were stained with Hoechst. Scale bars 50 μm. Results are expressed as mean ± SD from the indicated number (N) of independent experiments: *P < 0.05; ***P < 0.001.