Fig. 4.
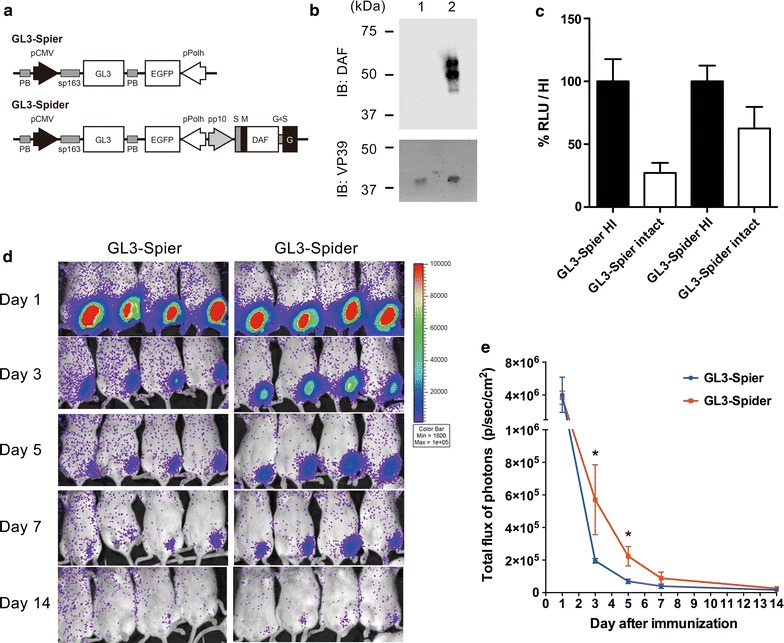
Schematic representation of the luciferase-expressing vaccines and their complement resistance in vitro and in vivo. a BES-GL3-Spier and BES-GL3-Spider vectors express firefly luciferase GL3 under the pCMV single promoter, and the BES-GL3-Spider vector contains a gene cassette for hDAF display. PB, PiggyBac transposon sequence; S, gp64 signal sequence; F, FLAG epitope tag; M, myc epitope tag; and EGFP, enhanced green fluorescent protein. b BES-GL3-Spier (lane 1) and BES-GL3-Spider (lane 2) were lysed and subjected to immunoblotting with anti-DAF and anti-VP39 Abs. c BES-GL3-Spier and BES-GL3-Spider were treated with heat-inactivated or intact serum and then transduced into HepG2 cells. Cell lysates were subjected to luciferase assays. Each relative luciferase unit (RLU) was normalized against each of the values of the heat-inactivated samples (n = 3). Bars and error bars indicate the mean ± SD of the values, respectively. Representative data from three independent studies are shown. HI, heat-inactivated serum; Intact, non-heat-inactivated serum. d Luciferase expression in BES-GL3-Spier- and BES-GL3-Spider-immunized Balb/c mice at 1, 3, 5, 7, and 14 days post-immunization (n = 4). The heat map image visible in the mice represents the total flux of photons (p/sec/cm2) in that area. e The mean total flux of photons is shown. Bars and error bars indicate the mean ± SD of the values, respectively. Comparison between groups was assessed by the Mann–Whitney U-test. *p < 0.05, compared with BES-GL3-Spier
