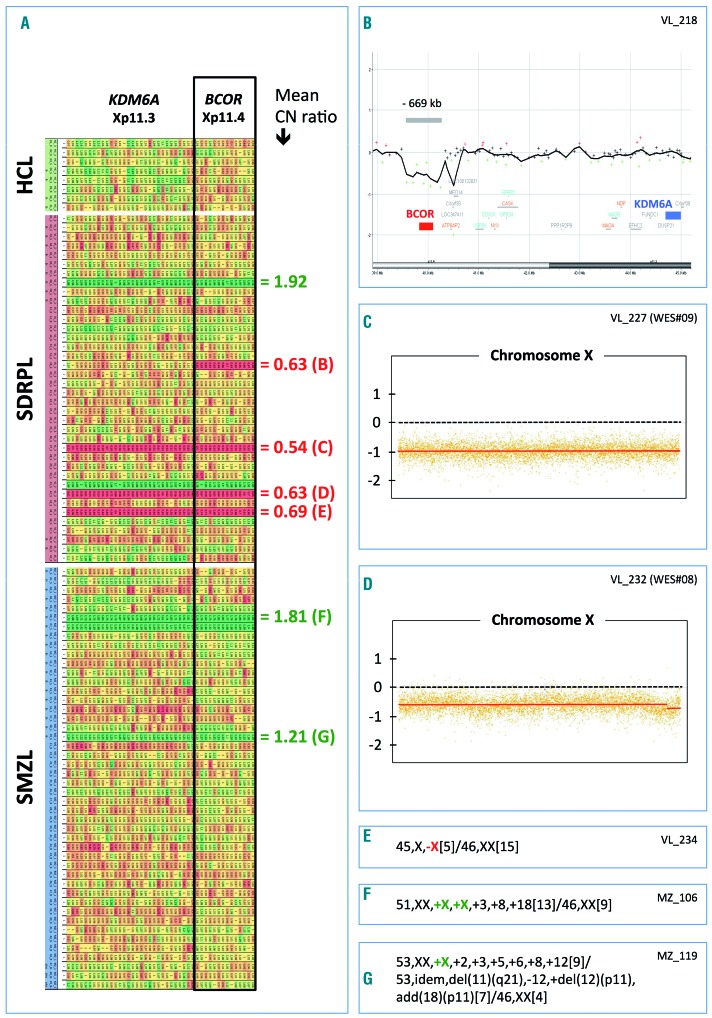
Figure 3.
Copy number evaluation at the BCOR Xp11.4 locus in hairy-cell leukemia, splenic diffuse red pulp lymphoma and splenic marginal zone lymphoma. (A) Copy number (CN) variation was detected from exon sequencing of the target genes after normalization using DeCovA.25,24 Each row represents a lymphoma case (HCL: hairy-cell lymphoma; SDRPL: splenic diffuse red pulp lymphoma; SMZL: splenic marginal zone lymphoma). Each column represents an exon of two different genes (BCOR and KDM6A) located at locus Xp11. A progressive gradation of CN variation distinguishes loss (red) and gain (green). Right: mean copy number ratio of the BCOR locus, with some references in brackets corresponding to the next inserts (B–G). (B) Microarray-based comparative genomic hybridization confirmed a monoallelic microdeletion of approximately 669 kb (arr[GRCh37] Xp11.4(39576746_40245183)×1), encompassing the BCOR locus (SDRPL female patient VL_218), whereas the KDM6A gene was unaffected. (C–D) Copy number variation of chromosome X detected by whole-exome sequencing using a circular binary segmentation algorithm and compared to a paired reference genome (log2 ratio). These two female patients with SDRPL acquired complete monosomy X. (E–G) Detailed karyotype results confirmed monosomy X (E), tetrasomy X (F) or trisomy X (G).