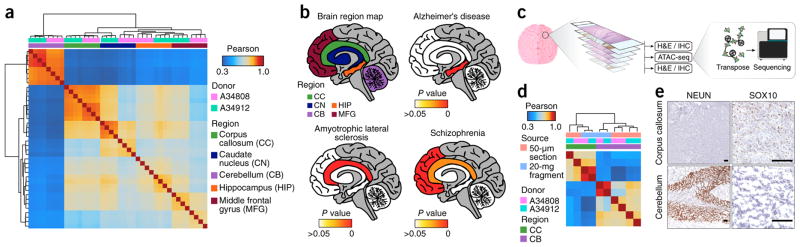
Figure 2.
Omni-ATAC in defined regions of the post-mortem human brain. (a) Pearson correlation heat map showing sample-by-sample unsupervised clustering on all peaks identified across all regions. Biological donor and brain region are indicated by the color across the top. CC, corpus callosum; CN, caudate nucleus; CB, cerebellum; HIP, hippocampus; MFG, middle frontal gyrus. All regions are represented by four technical replicates per donor. (b) Significance of enrichment of disease-specific GWAS polymorphisms in the uniquely accessible regions of five different brain regions (shown in the top left image) from individuals with the indicated diseases. The empirical P value is depicted colorimetrically with reference to association-based permutations of the GWAS SNPs. Nonsignificant empirical P values are represented in white. (c) Schematic of the experimental strategy for the generation of chromatin accessibility profiles from 50-μm frozen tissue sections with adjacent relevant histological staining. (d) Pearson correlation heat map showing sample-by-sample unsupervised clustering on all peaks. Data derived from the 20-mg tissue fragments represent the mean of four technical replicates from a single biological donor. Data derived from the 50-μm tissue sections show individual technical replicates. (e) Representative images (n = 3) showing histology and anti-NEUN (left) or anti-SOX10 (right) staining of 5-μm frozen sections from the corpus callosum (top) and cerebellum (bottom) immediately adjacent to the 50-μm section used for ATAC-seq shown in Supplementary Figure 8. Scale bars, 100 μm. SOX10 staining is shown at a higher resolution due to a more diffuse staining pattern.