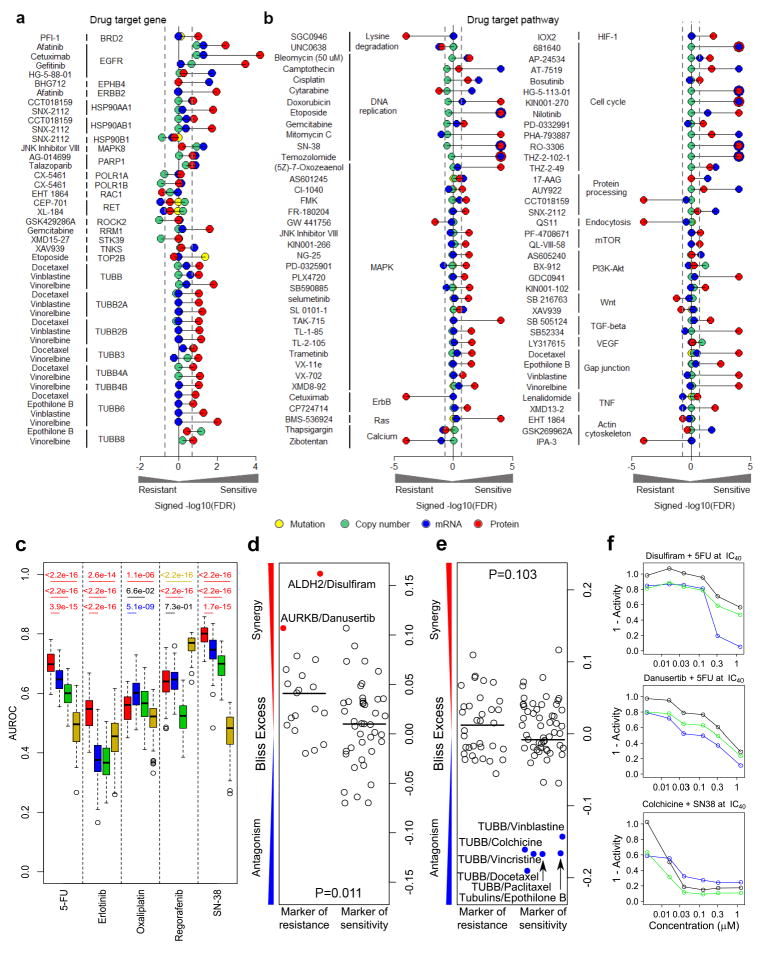
Figure 6. Proteomics data utility for predicting therapeutic responses.
(a, b) Associations of proteomic, mutation, DNA copy number and mRNA data with (a) established drug-target associations and (b) drug-pathway associations. Associations are shown for drug-target gene associations quantifiable at the protein level and significant in at least one of the four modalities as signed -log10(FDR) values from voom/limma and GSEA analyses, respectively. (c) Comparison of the utility of four omic modalities to predict drug sensitivity for 5-fluoruracil (5-FU), erlotinib, oxaliplatin, regorafenib and SN-38: proteomic data (red); RNA-Seq data (blue); CNA data (green); and exome mutation data (yellow). For each drug-omic modality combination, area under the receiver operating characteristic curve (AUROCs) were generated from 100 times of 5-fold cross-validations. The two-sided Wilcoxon rank sum test was used to compare the performance between protein-based models and models based on other omics data types. For each comparison, the p value is colored based on the color of the omic data type with significantly better performance. (d–e) Pharmacological targeting of proteins associated with resistance or sensitivity to (d) 5-FU or (e) SN-38. Bliss excess values are shown for drug combinations with 5-FU (at IC30 concentration) and SN-38 (at IC40 concentration) in HCT116 cells. The protein targets were restricted to those with FDR< 0.2 from the relevant voom/limma calculation; drugs are detailed in Supplementary Tables 4–5. p-values for Student’s t-test. (f) Dose-response plots for selected compounds alone (black), with either a 5-FU or SN-38 (blue), or the predicted response under the assumption of Bliss independence for the two compounds (green). Bliss synergy = blue line below green line; Bliss antagonism = blue line above green line.