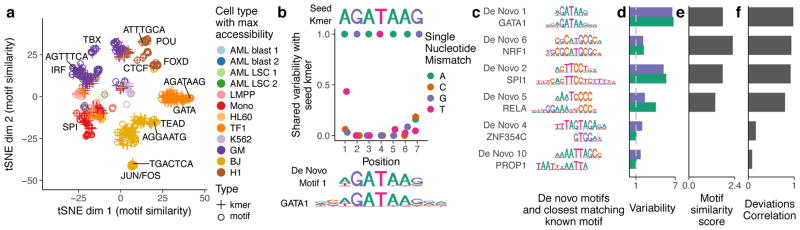
Figure 3. chromVAR identifies de novo motifs associated with chromatin accessibility variation in single cells.
(a) tSNE visualization of similarity between motifs and kmers based on the vector of normalized deviations across different cells. Labels highlight predominant families of motifs within a cluster and example kmers (b) For the seed kmer “AGATAAG”, the shared variability of k-mers with one mismatch from the seed kmer. The shared variability is defined as the square of the covariance of the deviation z-scores for the two kmers divided by the variance of the seed kmer for covariances greater than zero, and zero otherwise. These shared variabilities were used to assemble a de novo motif, shown under the plot along with the GATA1 motif. (c) Example de novo motifs assembled by chromVAR using deviations scores for 7-mers, along with the closest matching known motif below it. (d) Variability for both the de novo motif and the known motif for each pair in panel (c). (e) Motif similarity score (see methods) between the de novo motif and the known motifs in (c) (f) The Pearson correlation between the normalized deviations of the de novo motif and the known motif for each pair in (c).