Abstract
Introduction
This study assessed KRAS mutation detection and functional characteristics across 13 distinct technologies and assays available in clinical practice, in a blinded manner.
Methods
Five distinct KRAS-mutant cell lines were used to study five clinically relevant KRAS mutations: p.G12C, p.G12D, p.G12V, p.G13D and p.Q61H. 50 cell line admixtures with low (50 and 100) mutant KRAS allele copies at 20%, 10%, 5%, 1% and 0.5% frequency were processed using quantitative PCR (qPCR) (n=3), matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF) (n=2), next-generation sequencing (NGS) (n=6), digital PCR (n=1) and Sanger capillary sequencing (n=1) assays. Important performance differences were revealed, particularly assay sensitivity and turnaround time.
Results
Overall 406/728 data points across all 13 technologies were identified correctly. Successful genotyping of admixtures ranged from 0% (Sanger sequencing) to 100% (NGS). 5/6 NGS platforms reported similar allelic frequency for each sample. One NGS assay detected mutations down to a frequency of 0.5% and correctly identified all 56 samples (Oncomine Focus Assay, Thermo Fisher Scientific). One qPCR (Idylla, Biocartis) and MALDI-TOF (UltraSEEK, Agena Bioscience) assay identified 96% (all 100 copies and 23/25 at 50 copies input) and 92% (23/25 at 100 copies and 23/25 at 50 copies input) of samples, respectively. The digital PCR assay (KRAS PrimePCR ddPCR, Bio-Rad Laboratories) identified 60% (100 copies) and 52% (50 copies) of samples correctly. Turnaround time from sample to results ranged from ~2 hours (Idylla CE-IVD) to 2 days (TruSight Tumor 15 and Sentosa CE-IVD), to 2 weeks for certain NGS assays; the level of required expertise ranged from minimal (Idylla CE-IVD) to high for some technologies.
Discussion
This comprehensive parallel assessment used high molecular weight cell line DNA as a model system to address key questions for a laboratory when implementing routine KRAS testing. As most of the technologies are available for additional molecular biomarkers, this study may be informative for other applications.
Keywords: NSCLC, EGFR mutation, NGS, ddPCR, MALDITOF, qPCR, platform comparison
Key questions.
What is already known about this subject?
Diagnostic testing for biomarkers such as EGFR mutations is a well-established method of informing optimal treatment decisions for patients with non-small cell lung cancer (NSCLC). Testing for some other biomarkers in NSCLC, such as KRAS, is less well established, however is commonly employed to detect KRAS mutations in colorectal cancer. Therefore there exist many KRAS mutation detection methods that could aid clinical practice in the NSCLC setting.
What does this study add?
Due to the increase in the numbers of clinical trials focusing on patients with mutant NSCLC, there is a particular need to evaluate the testing options available. The aim of this study was to assess different diagnostic methods on how accurately they can identify KRAS mutations in samples characteristic of those used in the clinic.
How might this impact on clinical practice?
The results of this study aim to assist in the selection of the most appropriate technology for KRAS mutation detection, with special consideration for those tissues with low copy numbers and small sample sizes. It also elaborates on the shortcomings of each technology, allowing for a more informed clinical decision regarding which tests are most appropriate in which situations. Lastly, most technologies used for the detection of KRAS mutations can also be used for other biomarkers; thus, the results of this study are potentially applicable to other solid tumours where it is necessary to determine mutation status.
Introduction
Lung cancer is the most prevalent of all cancers, resulting in 1.38 million deaths every year,1 with over 80% being non-small cell lung cancer (NSCLC).2 3 Significant improvements in NSCLC treatment have been made with targeted tyrosine kinase inhibitors (TKIs), specifically those that target tumours with an EGFR mutation, including the EGFR T790M mutation, or ALK rearrangement.4–8 Patients needing access to these drugs must have a diagnostic test performed in order to ensure the correct drug is prescribed.
The recent approval of the T790M-directed EGFR-TKI, osimertinib,8 and the development of the MEK1/2 inhibitors, selumetinib (AZD6244, ARRY-142886),9 cobimetinib, trametinib and binimetinib, may further increase the demand for molecular characterisation of DNA derived from tumour tissue in patients with lung cancer.7 10 KRAS testing is commonly used to guide colorectal cancer (CRC) treatment; hence, there are a number of KRAS assays for testing laboratories to aid clinical practice; however, there is still a need for standardisation of testing methods, and uptake of optimal methods for routine diagnostics in NSCLC remains a challenge.11
Sanger capillary sequencing has long been the gold standard for DNA sequence analysis; however, it does not offer the high sensitivity required to detect somatic mutations at allelic frequencies less than ~20%.12 Numerous comparisons of technology platforms for EGFR 13 14 and KRAS (mainly in CRC15–23) mutation testing conclude that quantitative PCR (qPCR)-based methods offer the sensitivity, tissue economy and turnaround time required by physicians to guide treatment decisions. The advent of next-generation sequencing (NGS) platforms offers pathology laboratories an unparalleled insight into the cancer genome including de novo detection of variants as well as known actionable targets.24 However, there are important differences in sample handling and coverage of genes across the various NGS technologies along with increasing sequencing cost for the additional coverage (sequencing depth) necessary to detect low-level variants.25 26
There are challenges associated with mutation testing in fixed lung tissue samples. NSCLC samples typically generate exceptionally variable amounts of amplifiable DNA compared with more accessible tumour material such as CRC, ranging from ~10 copies/µL to tens of thousands of copies/µL.15 This reflects differences in accessibility of biopsy material and complexities in fixation due to lung tissue physiology. Even when sufficient quantities of DNA are obtained, not all is amplifiable and some quantification methods (eg, optical density and intercalating dyes) can overestimate the evaluable DNA for subsequent diagnostic testing.
The aim of the present study was to assess the ability of different mutation detection platforms to accurately identify low copy numbers of mutant KRAS DNA in varying backgrounds of wild-type DNA, and to combine this information with functional characteristics to aid in the assessment of a given testing platform.
Cell lines were chosen instead of formalin fixed paraffin embedded (FFPE) controls to ensure an adequate starting amount of DNA was available to produce identical samples for the high number of individual tests being evaluated. Some of the admixtures required large quantities of wild-type DNA not readily obtainable from FFPE. For example, the 0.5% mutant admixtures with 100 mutant copies required 19 900 copies/µL of wild-type (64 ng/µL total). Stability of smaller DNA fragments in FFPE tissue may have also presented issues in more dilute admixtures. It is acknowledged that FFPE tissue would represent clinical tissue more accurately; however, this was not deemed suitable due to the factors above.
Methods
Cell line models
Five distinct KRAS mutated cell lines, MIA PACA-2, PANC-1, MDA-MB231, SW620 and NCI-H460, were obtained from ATCC (Teddington, UK) (table 1). The KRAS mutations selected were p.G12C, p.G12D, p.G12V, p.G13D and p.Q61H, reflecting the most common KRAS variants according to codon associated with NSCLC. Cell lines were grown at the AstraZeneca cell bank (Alderley Park, UK), according to recommended conditions.
Table 1.
Characteristics of the five cell lines with known KRAS mutations
| Cell line | Base change | KRAS mutation | Amino acid change | Zygosity | ATCC order number | Reference |
| MIA PACA-2 | c.34G>T | p.Gly12Cys | 12C | Homozygous | CRL-1420 | (44) |
| PANC-1 | c.35G>A | p.Gly12Asp | 12D | Heterozygous | CRL-2547 | (45) |
| MDA-MB231 | c.38G>A | p.Gly13Asp | 13D | Heterozygous | HTB-26 | (46) |
| SW620 | c.35G>T | p.Gly12Val | 12V | Homozygous | CCL-227 | (47) |
| NCI-H460 | c.183A>T | p.Glu61His | 61H | Homozygous | HTB-177 | (48) |
DNA extraction and creation of admixtures
Cell line DNA was extracted from a frozen cell pellet containing ~5×106 cells using the Qiagen DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). KRAS mutation admixtures were created by quantifying in triplicate the high molecular weight DNA using a NanoDrop 8000 UV-Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Then, 1000 copies/µL DNA mutant standards were made from each cell line, which was diluted with known KRAS wild-type human genomic reference DNA (Roche Diagnostics, Mannheim, Germany) to create 10 distinct admixtures per mutation with mutant allele frequencies of 20%, 10%, 5%, 1% and 0.5% (table 2), assuming 50% allelic frequency with the heterozygous samples.
Table 2.
Relative copy numbers of mutant and wild-type KRAS DNA samples used to create the admixtures
| Sample number | Mutation (%) | Copies of mutant/μL | Copies of wild-type/μL |
| 1 | 20 | 100 | 400 |
| 2 | 10 | 100 | 900 |
| 3 | 5 | 100 | 1900 |
| 4 | 1 | 100 | 9900 |
| 5 | 0.5 | 100 | 19 900 |
| 6 | 20 | 50 | 200 |
| 7 | 10 | 50 | 450 |
| 8 | 5 | 50 | 950 |
| 9 | 1 | 50 | 4950 |
| 10 | 0.5 | 50 | 9950 |
Two sets of admixtures were created for all concentrations: one with 100 copies/µL mutant allele and one with 50 copies/µL mutant allele, resulting in a total series of 50 cell line admixtures. In addition to the 50 cell lines admixtures, six wild-type control samples were prepared with total input DNA copy numbers equivalent to those in the cell line admixtures. Admixtures were prepared in Axygen Maxymum recovery 1.7 mL tubes (Corning, Wiesbaden, Germany). The six wild-type controls contained 400, 1900, 9900 and 19 900 copies/µL, respectively.
Mother and daughter plate creation for distribution to study participants
Admixtures were created in multiwell plates as described in table 2. A plate schema was created where all 56 samples were randomly located across the plate. The samples were then transferred, witnessed by two scientists, to a DNase-free and RNase-free low retention ‘mother’ plate sufficient to perform the experiments. The material was frozen at −20°C until used. Daughter copies of the mother plate were then replicated using a Liquidator 96 (Mettler-Toledo, Columbus, Ohio, USA) manual pipetting system to minimise any transposition errors and were shipped on dry ice to the respective testing laboratories with instruction to keep frozen at −15 to −25°C until further processed. Instructions to thaw and thoroughly mix the samples prior to use using a vortex were given to all participants.
KRAS testing
KRAS mutation testing was carried out in a blinded manner on all 56 samples using 13 technologies and assays as outlined in online supplementary table 1. One test per sample was permitted unless part of an established repeat testing procedure (see online supplementary methods). KRAS mutation status was assessed using real-time PCR assays (therascreen KRAS RGQ PCR Kit, Qiagen; cobas KRAS Mutation Test, Roche Diagnostics; Idylla, Biocartis, Mechelen, Belgium); matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF) assays (iPLEX Pro tests, Agena Bioscience, San Diego, California, USA); NGS assays (ThunderBolts Cancer Panel, RainDance Technologies, Billerica, Massachusetts, USA; Oncomine Focus Assay, Thermo Fisher Scientific; Sentosa SQ NSCLC Panel, Vela Diagnostics, Singapore); Illumina Nextera Rapid Capture Custom Lung Panel, Cancer Research United Kingdom, London, UK; Ion AmpliSeq Cancer Hotspot Panel v2, Thermo Fisher Scientific; TruSight Tumor 15, Illumina, San Diego, California, USA; a droplet digital PCR (ddPCR) assay (PrimePCR ddPCR Mutation Assays KRAS, Bio-Rad Laboratories, San Diego, California, USA); and Sanger sequencing (Sanger capillary sequencing, Applied Biosystems, California, USA). Full details of the KRAS testing methodology are given in the online supplementary materials.
esmoopen-2017-000235supp001.pdf (512.1KB, pdf)
Results
Overall, analysis of the DNA admixtures using the 13 different technologies showed a range of performance for the different assays (table 3). Greater levels of mutation detection were achieved with the 100 copy DNA samples than with the 50 copy samples (figure 1A,B). The NGS results indicated that in some cases the actual mutation percentages of the admixtures differed considerably from the nominal. Therefore, the mutation percentages described below should be taken as nominal for comparison across assays. Of note, in clinical practice the percentage of mutant allele is used to estimate the limit of detection (LoD) of minor alleles during an assay validation. This would then be used to determine the minimal percentage of tumour cells in a specimen accepted to perform clinical testing and to interpret test results and would need to be established definitively. The LoD for each technology is shown in table 3.
Table 3.
KRAS mutation detection success by codon, concentration and technology
| (A) 100 mutant copies input | |||||||||||||
| Real-time quantitative PCR | MALDI-TOF | Next-generation sequencing | Droplet digital PCR |
Sanger capillary sequencing | |||||||||
| Nominal total copies of WT DNA | thera screen KRAS RGQ PCR Kit |
cobas KRAS Mutation Test | Idylla KRAS Mutation Test (point of care) | iPLEX Pro | UltraSEEK | Thunder Bolts |
Oncomine Focus Assay | Sentosa SQ NSCLC Panel | Ion AmpliSeq Cancer Hotspot Panel v2 | TruSight Tumour 15 | PrimePCR ddPCR Mutation Assays KRAS |
ABI3730 Sequencing | |
| p.G12C | 20 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | NMD | ✓ MD 14.3% | ✓ MD 20.5% | ✓ MD 11.1% | NMD | IMD | ✓ NMD |
| 10 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD 4.7% | ✓ MD 6.6% | ✓ MD 5.7% | ✓ MD 5.7% | ✓ MD 4.8% | ✓ MD | ✓ NMD | |
| 5 | ✓ MD | ✓ NMD | ✓ MD | ✓ MD | ✓ MD | ✓ MD 3.7% | ✓MD 2.4% | ✓ NMD 3.1% | ✓ MD 2.6% | ✓ MD 2.7% | ✓ MD | ✓ NMD | |
| 1 | ✓ NMD | ✓ NMD | ✓ MD | ✓ MD | ✓ MD | ✓ NMD | ✓ MD 0.5% | ✓ NMD– | ✓ NMD 0.0% | ✓ NMD 0.0% | ✓ MD | ✓ NMD | |
| 0.5 | ✓ NMD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD 0.3% | ✓ NMD– | ✓ NMD 0.0% | ✓ NMD 0.0% | NMD | ✓ NMD | |
| p.G12D | 20 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD 17.2% | ✓ MD 19.6% | ✓ MD 25.6% | ✓ MD 27.5% | ✓ MD 31.1% | ✓ MD | ✓ NMD |
| 10 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD 11.4% | ✓ MD 11.8% | ✓ MD 14.3% | ✓ MD 13.2% | NMD | ✓ MD | ✓ NMD | |
| 5 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD 7.6% | ✓ MD 5.8% | ✓ MD 6.7% | ✓ MD 5.1% | ✓ MD 7.3% | ✓ MD | ✓ NMD | |
| 1 | ✓ NMD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | ✓ MD 1.5% | ✓ MD 1.2% | ✓ NMD– | ✓ MD 1.0% | ✓ MD 1.6% | ✓ MD | ✓ NMD | |
| 0.5 | ✓ NMD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD 0.6% | ✓ NMD– | ✓ NMD 0.0% | ✓ NMD 0.0% | NMD | ✓ NMD | |
| p.G13D | 20 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | NMD | ✓ MD 13.9% | ✓ MD 14.6% | ✓ MD 10.4% | NMD | ✓ MD | ✓ NMD |
| 10 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD 2.7% | ✓ MD 7.4% | ✓ MD 7.7% | ✓ MD 8.6% | ✓ MD 6.4% | ✓ MD | ✓ NMD | |
| 5 | ✓ NMD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD 3.9% | ✓ MD 3.1% | ✓ MD 4.6% | ✓ MD 1.6% | ✓ MD 3.8% | ✓ MD | ✓ NMD | |
| 1 | ✓ NMD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD 0.9% | ✓ NMD– | ✓ NMD 0.0% | ✓ NMD 0.0% | ✓ MD | ✓ NMD | |
| 0.5 | ✓ NMD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | NMD | ✓ MD 0.4% | ✓ NMD– | ✓ NMD 0.0% | ✓ NMD 0.0% | NMD | ✓ NMD | |
| p.G12V | 20 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | NMD | ✓ MD 30.6% | ✓ MD 39.4% | ✓ MD 27.8% | NMD | ✓ MD | ✓ NMD |
| 10 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD 5.9% | ✓ MD 16.2% | ✓ MD 24.5% | ✓ MD 15.6% | ✓ MD 13.6% | IMD | ✓ NMD | |
| 5 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD 6.1% | ✓ MD 8.1% | ✓ MD 9.6% | ✓ MD 8.3% | ✓ MD 8.5% | ✓ MD | ✓ NMD | |
| 1 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ NMD | ✓ MD 1.3% | ✓ NMD 1.6% | ✓ MD 1.4% | ✓ MD 1.5% | ✓ MD | ✓ NMD | |
| 0.5 | ✓ MD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD 1.0% | ✓ NMD– | ✓ NMD 0.0% | ✓ NMD 0.0% | NMD | ✓ NMD | |
| p.Q61H | 20 | NA | ✓ MD | ✓ MD | ✓ MD | ✓ MD | NMD | ✓ MD 6.3% | ✓ MD 7.8% | ✓ MD 1.1% | NMD | IMD | ✓ NMD |
| 10 | NA | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | ✓ MD 2.3% | ✓ MD 2.7% | ✓ NMD 2.4% | ✓ MD 3.0% | ✓ MD 4.3% | IMD | ✓ NMD | |
| 5 | NA | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD 1.2% | ✓ NMD 2.5% | ✓ MD 1.8% | ✓ MD 1.5% | IMD | ✓ NMD | |
| 1 | NA | ✓ NMD | ✓ MD | ✓ NMD | ✓ NMD | ✓ NMD | ✓ MD 0.2% | ✓ NMD– | ✓ NMD 0.0% | ✓ NMD 0.0% | IMD | ✓ NMD | |
| 0.5 | NA | ✓ NMD | ✓ MD | ✓ NMD | ✓ NMD | ✓ NMD | ✓ MD 0.1% | ✓ NMD– | ✓ NMD 0.0% | ✓ NMD 0.0% | NMD | ✓ NMD | |
| (B) 50 mutant copies input | |||||||||||||
| Real-time quantitative PCR | MALDI-TOF | Next-generation sequencing | Droplet digital PCR |
Sanger capillary sequencing | |||||||||
| Nominal total copies of WT DNA | thera screen KRAS RGQ PCR Kit |
cobas KRAS Mutation Test | Idylla KRAS Mutation Test (point of care) | iPLEX Pro | UltraSEEK | Thunder Bolts |
Oncomine Focus Assay | Sentosa SQ NSCLC Panel | Ion AmpliSeq Cancer Hotspot Panel v2 | TruSight Tumor 15 | PrimePCR ddPCR Mutation Assays KRAS |
ABI3730 Sequencing | |
| p.G12C | 20 | NMD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | NMD | ✓ MD 10.8% | ✓ MD 19.8% | ✓ MD 13.1% | NMD | ✓ MD | ✓ NMD |
| 10 | ✓ MD | ✓ MD | ✓ MD | ✓ NMD | ✓ MD | NMD | ✓ MD 5.8% | ✓ MD 6.3% | ✓ MD 7.1% | NMD | ✓ MD | ✓ NMD | |
| 5 | ✓ NMD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | ✓ MD 2.1% | ✓ MD 2.9% | ✓ MD 3.3% | ✓ MD 1.9% | ✓ MD 3.6% | ✓ MD | ✓ NMD | |
| 1 | ✓ MD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD 0.6% | ✓ NMD - | ✓ NMD 0.0% | ✓ NMD 0.0% | ✓ MD | ✓ NMD | |
| 0.5 | ✓ NMD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD 0.4% | ✓ NMD− | ✓ NMD 0.0% | ✓ NMD 0.0% | IMD | ✓ NMD | |
| p.G12D | 20 | NMD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | NMD | ✓ MD 22.0% | ✓ MD 16.6% | ✓ MD 23.2% | NMD | ✓ MD | ✓ NMD |
| 10 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD 9.3% | ✓ MD 11.5% | ✓ MD 18.4% | ✓ MD 8.4% | ✓ MD 9.7% | ✓ MD | ✓ NMD | |
| 5 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD 5.0% | ✓ MD 5.0% | ✓ MD 5.2% | ✓ MD 7.6% | ✓ MD 6.7% | ✓ MD | ✓ NMD | |
| 1 | ✓ NMD | ✓ NMD | ✓ MD | ✓ MD | ✓ MD | ✓ NMD | ✓ MD 1.2% | ✓ NMD 2.2% | ✓ MD 1.9% | ✓ NMD 0.0% | ✓ MD | ✓ NMD | |
| 0.5 | ✓ NMD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD 0.3% | ✓ NMD - | ✓ MD 1.2% | ✓ NMD 0.0% | IMD | ✓ NMD | |
| p.G13D | 20 | NMD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | NMD | ✓ MD 13.0% | ✓ MD 6.5% | ✓ MD 15.5% | NMD | ✓ MD | ✓ NMD |
| 10 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | NMD | ✓ MD 6.6% | ✓ MD 6.6% | ✓ MD 6.1% | ✓ MD 2.7% | IMD | ✓ NMD | |
| 5 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD 2.8% | ✓ MD 2.9% | ✓ NMD 2.5% | ✓ MD 2.0% | ✓ MD 2.6% | ✓ MD | ✓ NMD | |
| 1 | ✓ NMD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD 0.8% | ✓ NMD - | ✓ NMD 0.0% | ✓ NMD 0.0% | ✓ MD | ✓ NMD | |
| 0.5 | ✓ NMD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD 0.4% | ✓ NMD - | ✓ NMD 0.0% | ✓ NMD 0.0% | IMD | ✓ NMD | |
| p.G12V | 20 | NMD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | NMD | ✓ MD 34.3% | ✓ MD 46.9% | ✓ MD 29.6% | ✓ MD 26.1% | ✓ MD | ✓ NMD |
| 10 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | NMD | ✓ MD 17.3% | ✓ MD 13.9% | ✓ MD 15.3% | NMD | IMD | ✓ NMD | |
| 5 | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD | ✓ MD 5.1% | ✓ MD 8.5% | ✓ MD 12.2% | ✓ MD 7.0% | ✓ MD 10.8% | IMD | ✓ NMD | |
| 1 | ✓ MD | ✓ NMD | ✓ MD | ✓ MD | ✓ MD | ✓ NMD | ✓ MD 1.7% | ✓ NMD− | ✓ MD 1.8% | ✓ MD 1.8% | IMD | ✓ NMD | |
| 0.5 | ✓ MD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD 0.8% | ✓ NMD− | ✓ NMD 0.0% | ✓ NMD 0.0% | ✓ MD | ✓ NMD | |
| p.Q61H | 20 | NA | ✓ MD | ✓ NMD | ✓ MD | ✓ MD | NMD | ✓ MD 6.0% | ✓ MD 10.3% | ✓ MD 4.5% | NMD | NMD | ✓ NMD |
| 10 | NA | ✓ MD | ✓ MD | ✓ MD | ✓ MD | NMD | ✓ MD 2.0% | ✓ NMD 2.8% | ✓ MD 3.7% | NMD | IMD | ✓ NMD | |
| 5 | NA | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD | ✓ NMD | ✓ MD 1.7% | ✓ NMD− | ✓ MD 1.6% | ✓ NMD 0.0% | IMD | ✓ NMD | |
| 1 | NA | ✓ NMD | ✓ NMD | ✓ NMD | ✓ NMD | ✓ NMD | ✓ MD 0.4% | ✓ NMD− | ✓ NMD 0.0% | ✓ NMD 0.0% | IMD | ✓ NMD | |
| 0.5 | NA | ✓ NMD | ✓ MD | ✓ NMD | ✓ NMD | ✓ NMD | ✓ MD 0.3% | ✓ NMD− | ✓ NMD 0.0% | ✓ NMD 0.0% | IMD | ✓ NMD | |
| (C) Wild-type only | |||||||||||||
| Real-time quantitative PCR | MALDI-TOF | Next-generation sequencing | Droplet digital PCR |
Sanger capillary sequencing | |||||||||
| Number of copies | thera screen KRAS RGQ PCR Kit |
cobas KRAS Mutation Test | Idylla KRAS Mutation Test (point of care) | iPLEX Pro | UltraSEEK | Thunder Bolts |
Oncomine Focus Assay | Sentosa SQ NSCLC Panel | Ion AmpliSeq Cancer Hotspot Panel v2 | TruSight Tumor 15 | PrimePCR ddPCR Mutation Assays KRAS |
ABI3730 Sequencing | |
| 20 000 | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | NMD | ✓ NMD | ✓WT | |
| 10 000 | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT 0.0% | NMD | ✓WT | |
| 2000 | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT 0.0% | NMD | ✓WT | |
| 1000 | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | NMD | ✓ NMD | ✓WT | |
| 500 | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | NMD | ✓WT | ✓WT | ✓WT | NMD | NMD | ✓WT | |
| 250 | ✓WT | ✓WT | ✓WT | ✓WT | ✓WT | NMD | ✓WT | ✓WT | ✓WT | NMD | NMD | ✓WT | |
✓MD, analysis successful, mutation detected; ✓NMD, analysis successful, but no mutation detected (in the case of the Sentosa assay, a mutation was detected but deemed to be below the defined cut-off); ✓WT, analysis successful, wild-type sample; NMD, analysis unsuccessful, no mutation detected; IMD, incorrect mutation detected; NA, kit does not assay codon.
MALDI-TOF, matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry.
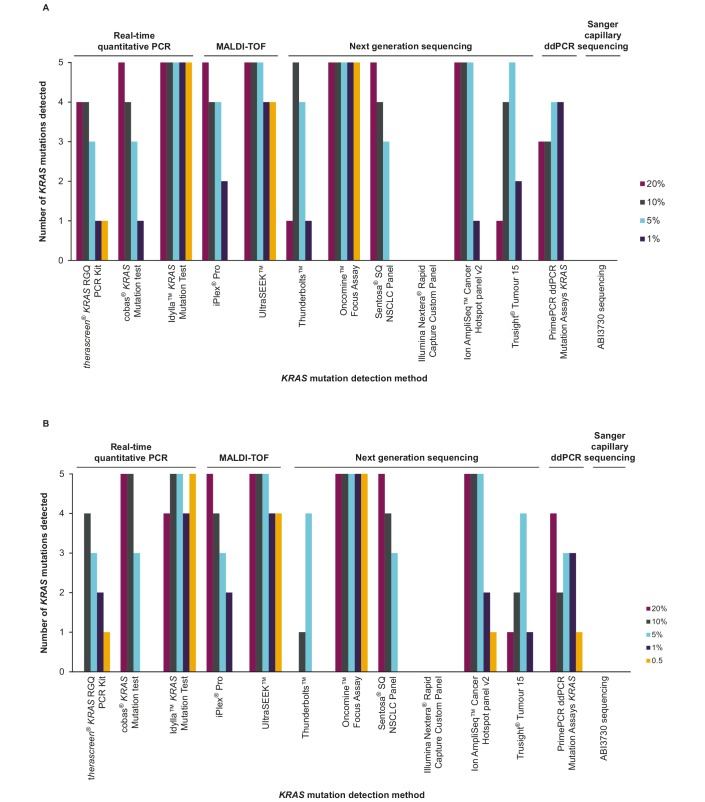
Figure 1.
(A) Number of KRAS mutations detected by mutation concentration by each technology with 100 copies mutant allele frequency. (B) Number of mutations detected by mutation concentration by each technology with 50 copies mutant allele frequency. ddPCR, droplet digital PCR; MALDI-TOF, matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry.
All technologies correctly identified all the wild-type control samples except the Nextera Rapid Capture Custom Lung Panel, ThunderBolts NGS assay and TruSight Tumor 15, where some or all samples were unsuccessfully analysed (see table 3C), but with no false-positive results being reported.
In the case of PrimePCR ddPCR Mutation Assays, four of the six wild-type samples were incorrectly identified as having mutations. Two of the four samples showed very low mutant frequencies of 0.08% G12C and 0.08% G12V in sample 1 and 0.2% G12D in sample 2, respectively, at high total numbers of positive droplets. Sample 3 showed a false-positive G12C mutation at a frequency of 1.1%. However, this sample had to be repeated as the first measurement did not deliver any result. This could indicate an assay performance problem in this sample. Sample 4 showed G12D/G12V mutations but at an extremely low total number of positive counts (<110 per assay). This again indicates a very low amount of DNA and data that are very difficult to interpret.
Real-time qPCR assays
In total, three qPCR-based assays were carried out. The therascreen KRAS RGQ PCR Kit was able to detect all codon 12 and 13 KRAS mutations at the 10% and 20% allele frequency level in the 100 copy sample, and three out of four of these mutations at the 5% level or lower. With the 50 copy samples, codons 12 and 13 mutations were detected only at the 10% level (table 3).
The cobas KRAS Mutation Test detected KRAS mutations successfully down to the 5% LoD, and lower for p.G12D, p.G13D and p.G12V in both the 100 copy and 50 copy samples. For p.G12C, the lowest level of detection was at the 10% level. For p.G12V mutations, cobas detected mutations as low as 1% in the 100 copy sample. The p.Q61H mutation was detected at 20% and 10% in the 100 and 50 copy samples, respectively.
The Idylla KRAS Mutation Test correctly identified the genotypes of KRAS p.G12C, p.G12D, p.G12V and p.G13D mutant samples at both mutant input levels. For KRAS p.Q61H, all mutant samples were identified at 100 copies input, while at 50 mutant copies input two samples expected to contain a p.Q61H mutation were scored as KRAS wild-type. Subsequent retesting after unblinding of these false-negative wild-type samples using Idylla KRAS multiplex PCR master mixes on a Bio-Rad CFX96 thermocycler identified one additional KRAS p.Q61H mutant sample, leaving only the 1% (50 copy) KRAS p.Q61H sample undetected. Overall, performance was thus in accordance with claimed LoD.
Matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry
The iPLEX Pro MALDI-TOF assay was able to detect all codon 12 and codon 13 KRAS mutations down to a level of 5% in the 100 and 50 copy number admixtures, with the exception of the p.G12C mutation which was only detected at the 20% level in the 50 copy number sample.
UltraSEEK detected all codon 12 and codon 13 mutations down to the 0.5% level in both the 100 copy and 50 copy samples. For the p.Q61H mutation, detection was achieved down to the 5% level with both the 50 and 100 copy samples.
Next-generation sequencing
We evaluated two different NGS library preparation principles, hybridisation capture and amplicon-based sequencing. The Illumina Nextera Rapid Capture Custom Lung Panel was not successful in identifying any KRAS mutants. After running the analysis, standard quality control filters were applied to eliminate poor quality calls (eg, due to issues such as stand bias and poor quality mapping score), and the resulting data did not detect any KRAS mutations. Relaxation of the quality control filters resulted in the detection of two common KRAS variants (p.G12D and p.G12C); however, the accuracy of these findings could not be determined as the genotype of the samples was not known at the outset of the study. Consequently, these results have not been included in further analyses.
Among the amplicon-based NGS tests, the Oncomine Focus Assay identified all five KRAS mutations studied. All mutations were detected in the 100 and 50 copy admixtures and at all levels of allele frequency down to 0.5%, the lowest level assessed (table 3).
The ThunderBolts NGS assay identified all codon 12 and 13 mutations. These KRAS mutations were detected at 5% with 100 copies and 50 copies of input, but not with all higher levels of allele frequency. For the p.Q61H mutation, this was detected at 10% with the 100 copy admixture.
The Sentosa SQ NSCLC Panel identified three out of four codon 12 and 13 mutations down to 5% and one down to 10% (p.G12C) with the 100 copy sample, and three out of four down to 5% with the 50 copy sample. It was able to quantify the allelic frequency of samples in cases that were below the established detection cut-off. For the p.Q61H mutation, the Sentosa panel detected only at the 20% level with both the 100 and 50 copy samples. The kit has a manufacturer-defined limit of 5% so that any results below 5% are defined as ‘mutation not detected’.
The Ion AmpliSeq Cancer Hotspot Panel v2 assay performed consistently well across both the 50 and 100 mutant allele copy admixtures. All five KRAS variants were detected in both cell admixtures, down to at least the 5% frequency. The assay also reported quantitative data and was able to detect mutations down to 1%.
The TruSight Tumor 15 assay identified all four codon 12 and 13 mutations to 5% and two of four to 1% (p.G12D and p.G12V) with the 100 copy sample. For the p.Q61H mutation, the TruSight Tumor 15 assay detected down to 5% frequency with the 100 copy sample. The assay identified all four codon 12 and 13 mutations to 5% and one of four to 1% (p.G12V) with the 50 copy sample. Some 10% and 20% levels of allele frequency were not identified due to nucleic acid input being below the manufacturer’s recommendation.
ddPCR assay
The PrimePCR ddPCR KRAS Mutation Assays were able to identify codon 12 and 13 mutations down to 1% with the 100 copy input. However, across both admixture and wild-type control samples, the assay identified the incorrect mutation in eight different mutation/allele frequency combinations (see table 3). The p.Q61H assay did not perform successfully in this experiment as none of the mutated samples were detected.
Sanger capillary sequencing
Sanger capillary sequencing produced weak PCR products and mutation peaks were only observable below the detection threshold. The overall results captured in table 3 show that this method did not identify any of the KRAS mutations at any of the levels of allele frequency tested.
Comparison of different assays
The detailed characteristics of the tested assays are reported in table 4, which provides an overview of the advantages of each assay, such as regulatory status provided. qPCR assays were observed to have the fastest turnaround time, with the Idylla KRAS Mutation Test being able to deliver the fastest turnaround time from sample to result a minimum of operator handling steps, while being a CE-marked product like therascreen and cobas tests. The UltraSEEK assay is able to detect low-level mutations with a quick turnaround time.
Table 4.
Mutation detection technology characteristics showing results of a questionnaire assessing handling, DNA input, sensitivity, turnaround time and multiplexing and regulatory status
| Real-time quantitative PCR | MALDI-TOF | Next-generation sequencing | Droplet digital PCR | Sanger capillary sequencing | |||||||||
| therascreen KRAS RGQ PCR Kit | cobas KRAS Mutation Test | Idylla KRAS Mutation Test | iPLEX Pro | UltraSEEK | ThunderBolts | Oncomine Focus Assay | Sentosa SQ NSCLC Panel | Illumina Nextera Rapid Capture Custom Lung Panel |
Ion AmpliSeq Cancer Hotspot Panel v2 | TruSight Tumor 15 panel |
PrimePCR ddPCR Mutation Assays KRAS | ABI3730 sequencing | |
| Ease of use | |||||||||||||
| Number of handling steps (wet work) | 6–8 | 3–5 | 1–2 | 3–5 | 3–5 | 11–20 | 6–8 | 6–8 | >20 | 11–20 | 6–8 | 6–8 | 11–20 |
| Number of handling steps (analysis to report) | 6–8 | 3–5 | 1–2 | 1–2 | 1–2 | 6–8 | 3–5 | 1–2 | 9–10 | 1–2 | 3–5 | 3–5 | 3–5 |
| Level of expertise required | 3 | 2 | 1 | 3 | 3 | 4 | 4 | 2 | 4 | 3 | 2 | 4 | 4 |
| Turnaround time | |||||||||||||
| Hands-on wet work time (excluding DNA extraction) | 2–5 hours | 31–60 min | 0–30 min | 31–60 min | 31–60 min | 5–10 hours | 5–10 hours | 2–5 hours | >20 hours | 10–20 hours | 10–20 hours | 2–5 hours | 2–5 hours |
| Hands-on analysis | 31–60 min | 0–30 min | 0–30 min | 0–30 min | 0–30 min | 1–2 hours | 31–60 min | 0–30 min | 2–5 hours | 1–2 hours | 0–30 min | 31–60 min | 2–5 hours |
| Total turnaround from DNA to clinical reporting (minimum) | 1–2 days | 2–4 hours | 2–4 hours | 1–2 days | 1–2 days | 5–10 days | 3–4 days | 2–3 days | 2–3 weeks | 5 days | 2 days | 1–2 days | 1–2 days |
| Multiplexing level | |||||||||||||
| Number of reactions per sample | 8+ | 2 | 3–5 | 1–24 | 1–4 | 2 | 1 | 1 | 1 | 1 | 2 | 3–5 | 2 |
| Maximum number of samples per run | 9 | 25–48 | 1 | 16–384 | 24–96 | 25–48 | 2–10 | 2–10 | 2–10 | 2–10 | 2–10 | 49–96 | 11–24 |
| Number of genes covered | 1 | 1 | 1 | 11+ | 11+ | 11+ | 11+ | 11+ | 11+ | 11+ | 11+ | 1 | 1 |
| Number of codons covered | 2 | 3 | 6–10 | 11+ | 11+ | Hot spots | Hot spots | Hot spots | Complete coverage | Complete coverage | Complete/ hot spot coverage |
3–5 | Complete coverage |
| Number of individual mutations | 6–10 | All possible | 11+ | 11+ | 11+ | All possible | All possible | All possible | All possible | All possible | All possible | 3–5 | All possible |
| Tissue/DNA requirements | |||||||||||||
| Minimum required DNA input | 20 ng | 50 ng | N/A | 80 ng | 10 ng | 20–30 ng | 10 ng | 5 ng | 50–75 ng | 10 ng | 10–20 ng | 1 ng | 10 ng |
| Amount of FFPE tissue required | 20 µm | 5 µm | 5–10 µm | 5 µm | 5 µm | 15–20 µm | 5 µm | 10 µm | 15–20 µm | 5 µm | 15–20 µm | 5 µm | 10 µm |
| Limit of detection | |||||||||||||
| Claimed sensitivity (%) | 5% | 5% | 5% | 10% | 0.1% | 5% | 5% | 5% | 10% | 5% | 5% | 0.001% | 20%–30% |
| Regulatory status | |||||||||||||
| Highest level of regulatory status achieved for assay | FDA-approved IVD CE-IVD |
FDA-approved IVD CE-IVD |
CE-IVD4 | Research use only | Research use only | Research use only | Research use only | CE-IVD | Research use only | Research use only | Research use only |
Research use only | Research use only |
CE-IVD, European Conformity- In-vitro Diagnostic; FDA, Food and Drug Administration; FFPE, Formalin Fixed Paraffin Embedded; IVD, In-vitro Diagnostic; MALDI-TOF, matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry; NSCLC, non-small cell lung cancer.
Typically, coverage beyond the usual mutation hot spots of the KRAS gene was achieved when using sequencing-based assays such as NGS or Sanger. The Idylla test and the cobas provided comprehensive coverage of the all the codons of interest in this study. For NGS, the KRAS mutation status was typically available in parallel with many other genes (at least ≥11; table 4). With respect to additional performance characteristics, NGS required more manual operating steps, with the exception of the CE-IVD Sentosa assay, which has a shorter turnaround time and provides a higher degree of automation. Sanger sequencing has comparable characteristics to the NGS technologies, with complete coverage and similar DNA requirements and throughput. Its coverage is limited to one gene and has low sensitivity.
Discussion
Here we aimed to establish a ‘snapshot’ of the relative performance of a range of currently established KRAS mutation detection platforms and assays. Our objective was to obtain information to assist in the selection of the most appropriate technology for KRAS mutation detection, particularly in those derived from small biopsies, which commonly have limited tissue available for testing. It is important to note that in some assays established DNA input requirements were exceeded or not met as part of this study and that this study does not show or claim to show superiority of one technology over another. In addition, the authors would like to point out the limitations of cell line admixtures to mimic clinical FFPE-derived samples and that validation using this sample type would be necessary for each technology in its laboratory setting.
The data showed that the use of admixture samples with low levels of DNA can result in variability in performance across the testing platforms and assays. Of the 13 assays evaluated in this work, nine showed relatively similar levels of accuracy and reliability in detecting KRAS mutations at low levels with varying sensitivities. Three assays, that is, Oncomine Focus Assay (NGS technology), Idylla KRAS Mutation Test (qPCR) and UltraSEEK (MALDI-TOF) performed particularly well, with high sensitivity and specificity across the entire cell line panel. The sensitivities of NGS, PCR and iPLEX Pro technologies were similar. In contrast, Sanger capillary sequencing failed to detect any mutations, as expected, due to its low sensitivity, which is the least of all the technologies, highlighting the need for optimisation for low copy DNA input levels.
Assay sensitivity is an important consideration with respect to the presence of tumour heterogeneity in clinical samples. The PrimePCR ddPCR assay generated some non-specific results, especially in the 50 mutant copy samples, which were concluded to be false-positive results due to the lower quantification limit being reached. We conclude that the sensitivity of the Bio-Rad ddPCR assays was not as high as proposed, especially not in the case of samples with low amounts of DNA. To optimise its further use, it would be recommended to establish positive controls with a known amount of mutant spike-in to wild-type samples at low levels.27 If such controls are used, a low-level mutant sample can be discriminated against a wild-type sample with very few false-positive mutant droplets and the lower limit of quantification can be lowered considerably.
Further, there are underlying reasons for some of these differences in performance; for example, only 3 of the 56 samples tested with the Illumina Nextera Rapid Capture Custom Lung Panel met the manufacturer’s recommended minimum DNA concentration of ≥50 ng/10 µL assay input, which may explain the apparent low sensitivity in the results. Additionally, the ThunderBolts Cancer Panel and the TruSight Tumor 15 assays recommended DNA concentration of ≥20 ng/10 µL, which may explain the lower sensitivity in the results. However, irrespective of the various reasons for the differences in performance observed in this analysis, the results highlighted that not all KRAS mutation assays were the same and the importance of choosing a mutation test that is appropriate for the quality and quantity of DNA in the sample type investigated.
Moreover, it is essential that the testing laboratory ensures appropriate validation of a mutation test with samples that are representative of the intended sample type has been carried out. Establishment of such tests is less laborious where the test is already validated and has regulatory approval, for example Food and Drug Administration (FDA) approval or CE-IVD certification. The Bio-Rad PrimePCR data in table 3 clearly show the potential issues of non-specific mutation detection when an assay has not been optimised appropriately for a given sample type. Commercially available ddPCR assays should be thoroughly optimised and validated before use.28 Reference standards can now be obtained for use to monitor assay performance, for example Horizon Diagnostics (Cambridge, UK) or AcroMetrix (Thermo Fisher Scientific). Where an incorrect test result could have a serious impact on patient health, external quality assessment schemes with ISO 17043 accreditation offer valuable support for molecular testing laboratories.
The NGS data showed that the p.G12V and p.Q61H mutation admixtures were almost double and almost half, respectively, of the intended nominal concentration. However, this is unlikely to affect the overall interpretation of the results of the study as the same samples were used consistently across all the technologies.
The admixture concentrations used in the analysis were designed to be challenging as many NSCLC patient samples or biopsies do not contain high levels of amplifiable DNA. Increasing use of fine needle aspirate (FNA), endobronchial ultrasound (EBUS)-guided biopsy and other small biopsy methods can result in challenging samples for the molecular pathology laboratory, with typically only 200–500 intact/evaluable cells per sample, and occasionally as few as 50 cells.29 30 Based on these data, we included a 50-copy admixture in addition to the 100-copy admixture combined with a wide range of wild-type background DNA.
We also demonstrated the KRAS mutation detection hit rates for the different technologies and may suggest the applicability of KRAS mutation tests based on the sensitivity data. However, it is important to also consider the range of sample types that may be encountered. In a clinical situation, only robust and thoroughly validated assays should be used. If a clinical laboratory tends to predominantly collect one or two sample types, then the choice of assay can be based on optimum performance with the expected quantitative DNA content for those samples, but an assay that is highly tolerant of sample variability without compromising assay performance should also be considered.
Our study had several limitations. Among them, the admixtures did not meet the recommended minimum DNA input requirements for the different technologies and assays in all cases, which may have affected subsequent template preparation or amplification and detection of KRAS mutations. However, the data generated show the comparative sensitivities various technologies can offer at the specified copy numbers, and can guide the selection of a test platform for use in a given application. Second, there may also be limitations to how the results can be extrapolated to performance on clinical samples. For instance, the failure rate for certain NGS mutation panels on FFPE samples can vary between 5% and 20%, depending on the technology used and on the implementation of preanalytical quality control measures.31 32 Further studies will be required to establish appropriate DNA input quantity cut-offs when assays are used with clinical FFPE-derived DNA, which contains many artefacts that can affect the DNA sequence.33 Third, extraction of the DNA from the cell lines was not performed in all instances with the validated method or kit specified for use with a given assay or workflow.
When choosing an appropriate platform and assay to meet the needs of a particular clinic laboratory, there are many factors to consider. In addition to the sensitivity of the assay being appropriate for the DNA content of the sample, it is also important to consider which KRAS mutations an assay can detect. For example, while the therascreen is an FDA-approved In-vitro Diagnostic (IVD) and European Conformity In-vitro Diagnostic (CE-IVD)-approved assay,34 it does not cover KRAS p.Q61 codon mutation, which accounts for up to 5% of KRAS mutations in NSCLC.15 In this case, the laboratory may need to consider if an additional KRAS mutation test should be used so that a larger number of hot spot mutations are tested for. Complete coverage of all clinically relevant codons in the KRAS gene is typically seen when using sequencing-based assays such as NGS or Sanger, or in case of large-panel qPCR assays such as in the Idylla test, or to lesser extent the cobas test. A clear advantage of NGS is that the KRAS mutation status is available in parallel with information on many other genes, making more use of precious limited amount of tumour material.
Furthermore, in order to assess the suitability of a given test for use in the clinic, other parameters associated with assay use need to be considered, such as ease of use, number of manual operating steps, turnaround time from clinical sample to result and the number of samples that can be assessed in one instrument run. Using an assay with a long turnaround time may not be appropriate when servicing a clinic sending high numbers of samples for mutation testing, or an assay requiring high levels of technical laboratory expertise may not be appropriate in a situation with limited resources. As shown in table 4, different assays have various characteristics that may be taken into consideration. For example, the CE-IVD Idylla KRAS Mutation Test automates the complete workflow after sectioning of the FFPE sample to result, and no manual pipetting or analysis steps are required to generate the test result, whereas the NGS Sentosa SQ NSCLC Panel assay (Vela Diagnostics), which is also a CE-IVD test, has a longer turnaround time but instead covers more target genes and analysis is automated. In general other NGS assays required more manual operating steps and somewhat more interpretation than the single-gene assays included in our study.
In conclusion, the variation between technologies and assays highlighted the need to select the most appropriate test for the particular clinical situation and the importance of appropriate test validation. Most technologies used to detect KRAS mutations are also available for other molecular biomarkers, meaning the results of this study may be applicable to other solid tumours where mutation status is needed for therapy selection.
Acknowledgments
This study was sponsored and supported by AstraZeneca. We thank Andrew Brooks of Bioprocessing Solutions Alliance for the RainDance ThunderBolts NGS data. Medical editing support was provided by Sandra Brave, PhD, of iMed Comms, an Ashfield Company, part of UDG Healthcare funded by AstraZeneca.
Footnotes
Contributors: Conceived and designed the experiments: JLS, HB, AK. Performed the experiments: JLS, AR, AS, GC, BC, BA, RC, BSGK, AOHN, ILD and PC. Analysed the data: JLS, AR, AS, GC, BC, BA, RC, BSGK and PC. Wrote the paper: JLS, AR, AS, GC, BC, BA, RC, BSGK, AOHN, ILD and PC.
Funding: AstraZeneca provided funds for this research.
Competing interests: JLS, HB and AK are employees and shareholders of AstraZeneca. AR is an employee of Birmingham Women’s NHS Foundation Trust. AS is an employee of IMGM Laboratories. GC is an employee of University of Cambridge. BC is an employee of Biocartis NV and has a patent pending for the isolation of nucleic acids. BA is an employee of Vela Diagnostics and Eppendorf. RC is an employee of NewGene. BSGK and PC are employees of Thermo Fisher Scientific. AOHN is a paid employee of Agena Bioscience and holds company stock options. ILD is an employee of Illumina and holds stock in the company, and an employee of Invivoscribe.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Ferlay J, Shin HR, Bray F, et al. . Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893–917. 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Siegel R, Ward E, et al. . Cancer statistics, 2009. CA Cancer J Clin 2009;59:225–49. 10.3322/caac.20006 [DOI] [PubMed] [Google Scholar]
- 3. Youlden DR, Cramb SM, Baade PD. The international epidemiology of lung cancer: geographical distribution and secular trends. J Thorac Oncol 2008;3:819–31. 10.1097/JTO.0b013e31818020eb [DOI] [PubMed] [Google Scholar]
- 4. Lindeman NI, Cagle PT, Beasley MB, et al. . Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol 2013;8:823–59. 10.1097/JTO.0b013e318290868f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lynch TJ, Bell DW, Sordella R, et al. . Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129–39. 10.1056/NEJMoa040938 [DOI] [PubMed] [Google Scholar]
- 6. Kris MG, Natale RB, Herbst RS, et al. . Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA 2003;290:2149–58. 10.1001/jama.290.16.2149 [DOI] [PubMed] [Google Scholar]
- 7. Pirker R. Novel drugs against non-small-cell lung cancer. Curr Opin Oncol 2014;26:145–51. 10.1097/CCO.0000000000000056 [DOI] [PubMed] [Google Scholar]
- 8. Jänne PA, Yang JC, Kim DW, et al. . AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689–99. 10.1056/NEJMoa1411817 [DOI] [PubMed] [Google Scholar]
- 9. Jänne PA, Shaw AT, Pereira JR, et al. . Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol 2013;14:38–47. 10.1016/S1470-2045(12)70489-8 [DOI] [PubMed] [Google Scholar]
- 10. Pirker R, Filipits M. Targeted therapies in lung cancer. Curr Pharm Des 2009;15:188–206. 10.2174/138161209787002915 [DOI] [PubMed] [Google Scholar]
- 11. Tan C, Du X. KRAS mutation testing in metastatic colorectal cancer. World J Gastroenterol 2012;18:5171–80. 10.3748/wjg.v18.i37.5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ellison G, Donald E, McWalter G, et al. . A comparison of ARMS and DNA sequencing for mutation analysis in clinical biopsy samples. J Exp Clin Cancer Res 2010;29:132 10.1186/1756-9966-29-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ellison G, Zhu G, Moulis A, et al. . EGFR mutation testing in lung cancer: a review of available methods and their use for analysis of tumour tissue and cytology samples. J Clin Pathol 2013;66:79–89. 10.1136/jclinpath-2012-201194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Penzel R, Sers C, Chen Y, et al. . EGFR mutation detection in NSCLC--assessment of diagnostic application and recommendations of the German Panel for Mutation Testing in NSCLC. Virchows Arch 2011;458:95–8. 10.1007/s00428-010-1000-y [DOI] [PubMed] [Google Scholar]
- 15. Sherwood JL, Müller S, Orr MC, et al. . Panel based MALDI-TOF tumour profiling is a sensitive method for detecting mutations in clinical non small cell lung cancer tumour. PLoS One 2014;9:e100566 10.1371/journal.pone.0100566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mosko MJ, Nakorchevsky AA, Flores E, et al. . Ultrasensitive detection of multiplexed somatic mutations using MALDI-TOF mass spectrometry. J Mol Diagn 2016;18:23–31. 10.1016/j.jmoldx.2015.08.001 [DOI] [PubMed] [Google Scholar]
- 17. Pinto P, Rocha P, Veiga I, et al. . Comparison of methodologies for KRAS mutation detection in metastatic colorectal cancer. Cancer Genet 2011;204:439–46. 10.1016/j.cancergen.2011.07.003 [DOI] [PubMed] [Google Scholar]
- 18. Lee S, Brophy VH, Cao J, et al. . Analytical performance of a PCR assay for the detection of KRAS mutations (codons 12/13 and 61) in formalin-fixed paraffin-embedded tissue samples of colorectal carcinoma. Virchows Arch 2012;460:141–9. 10.1007/s00428-011-1180-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Domagała P, Hybiak J, Sulżyc-Bielicka V, et al. . KRAS mutation testing in colorectal cancer as an example of the pathologist’s role in personalized targeted therapy: a practical approach. Pol J Pathol 2012;63:145–64. [DOI] [PubMed] [Google Scholar]
- 20. Bolton L, Reiman A, Lucas K, et al. . KRAS mutation analysis by PCR: a comparison of two methods. PLoS One 2015;10:e0115672 10.1371/journal.pone.0115672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Franklin WA, Haney J, Sugita M, et al. . KRAS mutation: comparison of testing methods and tissue sampling techniques in colon cancer. J Mol Diagn 2010;12:43–50. 10.2353/jmoldx.2010.080131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gonzalez de Castro D, Angulo B, Gomez B, et al. . A comparison of three methods for detecting KRAS mutations in formalin-fixed colorectal cancer specimens. Br J Cancer 2012;107:345–51. 10.1038/bjc.2012.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kimura H, Ohira T, Uchida O, et al. . Analytical performance of the cobas EGFR mutation assay for Japanese non-small-cell lung cancer. Lung Cancer 2014;83:329–33. 10.1016/j.lungcan.2013.12.012 [DOI] [PubMed] [Google Scholar]
- 24. Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet 2008;24:133–41. 10.1016/j.tig.2007.12.007 [DOI] [PubMed] [Google Scholar]
- 25. Quail MA, Smith M, Coupland P, et al. . A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 2012;13:341 10.1186/1471-2164-13-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu L, Li Y, Li S, et al. . Comparison of next-generation sequencing systems. J Biomed Biotechnol 2012;2012:1–11. 10.1155/2012/251364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sacher AG, Paweletz C, Dahlberg SE, et al. . Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol 2016;2:1014–22. 10.1001/jamaoncol.2016.0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pender A, Garcia-Murillas I, Rana S, et al. . Efficient Genotyping of KRAS Mutant Non-Small Cell Lung Cancer Using a Multiplexed Droplet Digital PCR Approach. PLoS One 2015;10:e0139074 10.1371/journal.pone.0139074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pirker R, Herth FJ, Kerr KM, et al. . Consensus for EGFR mutation testing in non-small cell lung cancer: results from a European workshop. J Thorac Oncol 2010;5:1706–13. 10.1097/JTO.0b013e3181f1c8de [DOI] [PubMed] [Google Scholar]
- 30. Thunnissen E, Kerr KM, Herth FJ, et al. . The challenge of NSCLC diagnosis and predictive analysis on small samples. Practical approach of a working group. Lung Cancer 2012;76:1–18. 10.1016/j.lungcan.2011.10.017 [DOI] [PubMed] [Google Scholar]
- 31. Al-Kateb H, Nguyen TT, Steger-May K, et al. . Identification of major factors associated with failed clinical molecular oncology testing performed by next generation sequencing (NGS). Mol Oncol 2015;9:1737–43. 10.1016/j.molonc.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goswami RS, Luthra R, Singh RR, et al. . Identification of Factors Affecting the Success of Next-Generation Sequencing Testing in Solid Tumors. Am J Clin Pathol 2016;145:222–37. 10.1093/ajcp/aqv023 [DOI] [PubMed] [Google Scholar]
- 33. Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem 2015;61:64–71. 10.1373/clinchem.2014.223040 [DOI] [PubMed] [Google Scholar]
- 34. Qiagen. Therascreen KRAS RGQ PCR kit handbook manchester. 2016. http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/DeviceApprovalsandClearances/Recently-ApprovedDevices/ucm400569.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
esmoopen-2017-000235supp001.pdf (512.1KB, pdf)