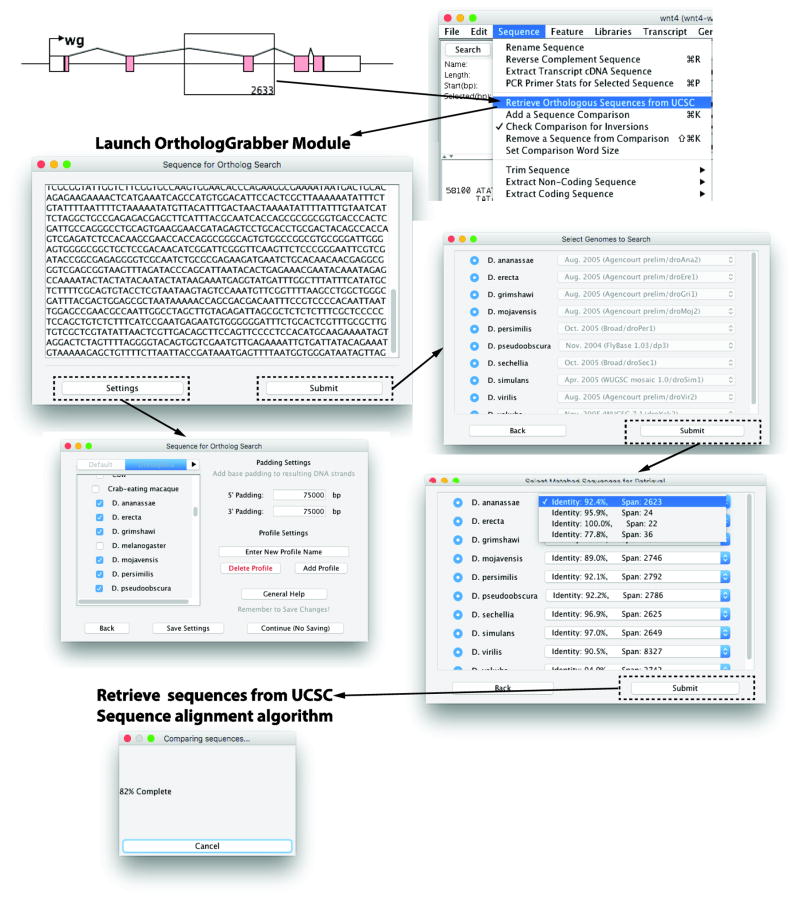
Figure 2. The OrthologGrabber module automates the retrieval of orthologous sequences from the UCSC genome database.
In the Graphical View, a user can click and drag a box to select a subregion of the currently loaded sequence to compare to the UCSC genome database. The OrthologGrabber module is launched through a menu item, opening a second java window. The Settings button allows the user to select the species to which the selected sequence will be compared, and specify the amount of flanking sequence to be retrieved from the database. Settings can be saved for frequent usage. Once the Submit button is pressed, the user is given the option to select among the different available versions of the selected genomes. The next window shows the results from the BLAT search, and users may select which BLAT match will be used for sequence retrieval. Retrieved sequences, including the specified amount of flanking sequence, are then fed back into GenePalette’s sequence alignment algorithm.