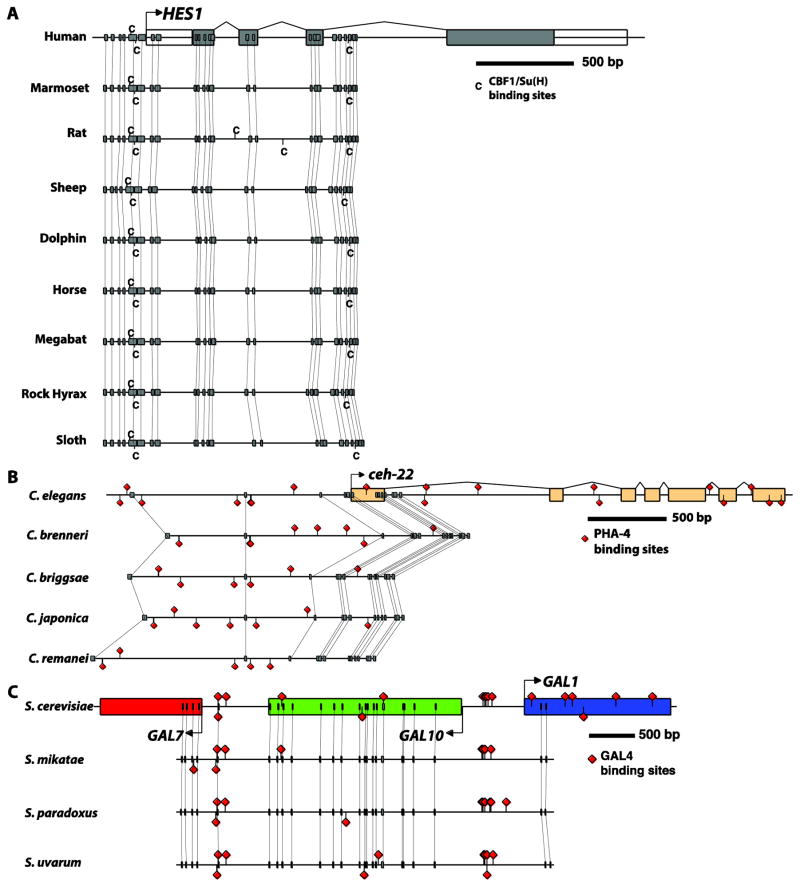
Figure 5. Examples of anchor-point assisted phylogenetic footprinting in multiple species.
(A) A highly conserved pair of binding sites for CBF1/Su(H) upstream of the vertebrate Hes1 gene, as reported in (Bailey and Posakony, 1995; Rebeiz et al., 2012). (B) Comparative analysis of PHA-4 binding sites that were described upstream of the C. elegans ceh-22 gene (Gaudet and Mango, 2002) reveals their rapid turnover. (C) Conservation of UAS sequences bound by the GAL4 protein (West et al., 1984) in the divergent upstream region of the GAL1 and GAL10 genes. GenePalette was used to download and align the schematized sequences. A Postscript file exported from GenePalette was modified in Adobe Illustrator to enhance the presentation. Yeast genomes represented in (C) were manually obtained through the synteny viewer from SGD (http://www.yeastgenome.org/cgi-bin/FUNGI/FungiMap).