Abstract
The hypothesis is discussed that prostate cancer marker lncRNA PCA3 was introduced into the human genome by an oncogenic virus, and that viral infection‐related mechanisms might underlie its overexpression and prostate cancer initiation and/or progression.

Subject Categories: Cancer; Microbiology, Virology & Host Pathogen Interaction; Urogenital System
Prostate cancer, the most frequent type of cancer in men, is surpassed only by lung cancer in causing cancer‐related death. Despite major progress in defining the mutational landscape of this tumor, its etiology remains obscure. The intronic long‐noncoding RNA (lncRNA) PCA3 is a specific marker of prostate cancer that acts as a trans‐dominant negative oncogene to down‐regulate the tumor suppressor gene PRUNE2. The unusual genomic organization and sequence of PCA3 leads us to hypothesize that it was introduced into the human genome by an as‐yet undefined oncogenic virus. We further suggest viral infection‐related mechanisms as functional in PCA3 overexpression and involved in prostate cancer initiation and/or progression. This is supported by known links between viral infections and prostate cancer, and the lack of this tumor in other mammals despite its high incidence in humans. Finally, we suggest that genomic sequence‐based approaches might help us uncover the potential role of viruses in prostate cancer etiology.
Introduction: The curious case of PCA3 in prostate cancer
The intronic lncRNA Prostate Cancer Antigen 3 (PCA3) is a highly specific biomarker: While only present at low levels in normal or benign hyperplastic prostate, its expression is increased up to 100‐fold in about 95% of prostatic carcinomas (Schalken et al, 2003). Despite this strong correlation, the biological function of PCA3 remained elusive for almost two decades after its discovery (Bussemakers et al, 1999), until we established its role as a trans‐dominant negative oncogene that downregulates a protein‐coding tumor suppressor gene, prune homolog 2 (PRUNE2) (Salameh et al, 2015). In experimental models of human prostate cancer, PCA3 overexpression as well as PRUNE2 silencing accelerates the growth of tumor xenografts in vivo, while increasing cell proliferation, adhesion, and migration in vitro (Salameh et al, 2015). In prostate cancer patients, decreased levels of PRUNE2 are indicative of poor prognosis, as this is associated with metastatic disease (National Center for Biotechnology Information, Gene Expression Omnibus, NCBI GEO, profile 34856174; n = 171) and a high Gleason score (The Cancer Genome Atlas, TCGA; n = 497). Surprisingly, data retrieved from large‐scale databases of human tissue samples (TCGA; and The Atlas of noncoding RNAs in Cancer, TANRIC) show homogeneous expression of PCA3 only in prostate cancer, but not in other tumor types, including those in which PRUNE2 loss‐of‐function events have been reported, that is, Merkel cell carcinoma, neuroblastoma, glioblastoma, and parathyroid carcinoma.
From an evolutionary standpoint, PCA3 is a relatively new gene: All four exons and the upstream (putative promoter‐containing) sequence are conserved among primates, but only exon 4 shares conservation with the rabbit (68% identity) and mouse (52% identity) orthologs. Prostate cancer occurrence is even more stringent than PCA3 conservation: Despite its high incidence in humans, it is rare or absent in other mammals (De Marzo et al, 1999), including nonhuman primates kept in captivity (ruling out the possibility that a shorter lifespan might account for the difference). Dogs are the only known exception, but even here the incidence is much lower than in humans. Moreover, while the vast majority of prostate cancers in men are androgen‐dependent adenocarcinomas, dogs more often present with androgen‐independent ductal carcinomas. A simple link between human diet and predisposition to prostate cancer (De Marzo et al, 1999) also does not satisfactorily explain the paradoxical (non)‐diffusion of this tumor. The intriguing association between PCA3 and prostatic carcinoma in human males is at odds with the established concept that oncogenes are shared among different tumor types and/or animal species. How might this tissue‐ and species‐specificity be linked to the currently unknown triggering event(s) in prostate cancer?
A connection between oncoviruses, long noncoding RNAs, and cancer: coevolution to optimize fitness
Tumor‐related lncRNA transcripts are often encoded and/or regulated by viruses. Indeed, all known oncoviruses, including hepatitis B and C viruses (HBV and HCV, respectively), Epstein–Barr virus (EBV), human papillomavirus (HPV), and Kaposi's sarcoma‐associated herpesvirus (KSHV), produce their own lncRNAs and/or induce the transcription of cellular lncRNAs, contributing to cancer predisposition (Li et al, 2016). This “virus‐lncRNA‐cancer” leitmotif has been validated in different malignant settings including HBV‐Highly Upregulated in Liver Cancer (HULC) in hepatocellular carcinoma, EBV‐Small Nucleolar RNA Host Gene 8 (SNHG8) in gastric cancer, and HPV‐HOX transcript antisense intergenic RNA (HOTAIR) in cervical cancer. The connection is not limited to ongoing infections: Ancient viruses and their animal hosts have actively exchanged portions of genetic material throughout evolution (Krupovic & Koonin, 2017). This phenomenon is related to the need of viruses to optimize their replication by exploiting the eukaryotic cell machinery. In addition, random viral insertions can lead to the formation of virus‐host chimeric functional units (e.g., lncRNAs) that are selected for their capacity to favor host functions in physiological and/or pathological conditions (e.g., cancer, inflammation, immunity). A consequence of this coevolution, the conservation between viral sequences and lncRNAs in at least mammals and birds, is beginning to be unraveled with regard to genomic structure and regulation. An interesting example in the poultry industry is the integration of several related avian leukosis viruses with miRNAs and lncRNAs to cause tumors, decreased fertility, and premature death (Lan et al, 2017).
Is PCA3 an oncovirus‐derived gene?
Several attributes of PCA3 suggest that this lncRNA may be derived from and/or regulated by viruses. First, the PCA3 promoter and exon 1 are included in a partially conserved long interspersed nuclear element type 2 (LINE‐2) repeat, which is an ancient virus‐derived retrotransposon. This is similar to the HBV‐regulated lncRNA HULC, whose promoter and first exon are embedded in a long‐terminal repeat (LTR) retrotransposon‐like sequence (Kapusta et al, 2013). Second, as would be expected if the PCA3 promoter were derived from a virus, it does not contain canonical transcription factor binding sites (e.g., TATA box) and shows no sequence conservation with any annotated human promoter. Third, PCA3 is located on the opposite DNA strand to intron 6 of the PRUNE2 gene. This configuration is reminiscent of the bidirectional transcription of the EBV latency origin of replication, with leftward transcripts being mainly noncoding and regulatory, and rightward transcripts containing open reading frames (Cao et al, 2015). Fourth, PRUNE2 downregulation by PCA3 occurs through adenosine deaminase acting on RNA (ADAR)‐mediated editing, a post‐transcriptional mechanism largely employed in the cellular response to viruses such as polyomavirus and EBV (Cao et al, 2015), and upregulated in EBV‐infected lymphocytes (http://www.gtexportal.org/home/gene/ADAR).
A tale of sex, viral transmission, and human prostate cancer
Human prostate cancer has been associated with several viruses. Digital expression studies have shown that the human endogenous retrovirus K (HERV‐H), a remnant of ancient infections fixed in the ancestral human genome 30–70 millions of years ago, is overexpressed in human stomach, colon and prostate cancers. The activation of an analogous retrovirus, HERV‐K, induces the production of corresponding antigens and immune responses (Reis et al, 2013) that correlate with prostate cancer progression. A few additional indications of a direct viral etiology also exist. The BK polyomavirus (BKV) seems to be a cofactor in early stages of prostate cancer (Das et al, 2008), and its correlation with malignant progression has been proposed. Overall, a 19% prevalence of polyomaviruses such as BKV, JC virus (JCV), and SV40 has been reported in prostate cancer cases, suggesting that the subclinical persistent infections that are frequent in the human population might contribute to the onset and/or progression of this malignancy. Positive association with other oncoviruses such as HPV has also been established. In general, an increased relative risk estimate for prostate cancer has been associated with sexually transmitted diseases (e.g., gonorrhea and syphilis) (Dennis & Dawson, 2002), supporting the possibility of infectious components in tumor development. Despite their limitations, these data are encouraging: The challenge of finding a direct link between viral infection and cancer is common to a number of pathologies for which standard causation rules, for example, Koch's postulates and Hill's criteria of causality (reviewed in Moore & Chang, 2014), do not apply. Heterogeneous etiologies have been described for other virus‐linked cancers, for example, Merkel cell carcinoma, where the presence of polyomavirus alone is not sufficient for tumor development as well as squamous cell carcinomas of the genital and head‐and‐neck regions, in which HPV infection appears to be a necessary yet insufficient cause (zur Hausen, 2002).
Conclusion: How could this hypothesis be tested?
To actually demonstrate the connection between viral infection, lncRNAs, and prostate cancer, the characterization of the variety of viruses present in tumor cells will be needed. Digital transcript subtraction has been successfully applied to uncover the association between polyomavirus and human Merkel cell carcinoma (Feng et al, 2008) and could also be employed to hunt for new oncoviruses in prostate cancer. To cover the full spectrum of virus‐related mechanisms, a properly designed variant of this in silico strategy should be designed to include noncoding RNAs. Such next‐generation approaches would rule out potential contaminations, as in the unfortunate precedent of xenotropic murine leukemia‐related virus (XMRV), a putative retrovirus involved in prostate cancer later demonstrated to be a laboratory artifact (Paprotka et al, 2011). Also, a systematic analysis of virus‐derived genomic sequences in a large cohort of prostate cancer patients would elucidate the structural connections between lncRNAs and ancient infections, as a preliminary step toward a comprehensive functional characterization of the corresponding mechanisms. Above all, this analysis should allow low stringency homology searches, to include poorly conserved sequences (such as the LINE‐2 repeat in which PCA3 is embedded) derived from millions of years of evolutionary pressure, and thereby barely recognizable.
In summary, we hypothesize that the prostate‐specific lncRNA PCA3 has been introduced into the human genome by an ancient virus, and that it is regulated by virus‐specific mechanisms (Fig 1). The identification of cellular and/or viral regulatory processes that are specifically engaged in prostate pathophysiology will be pivotal for understanding the contribution of PCA3 and other noncoding RNA transcripts in the onset and progression of prostate cancer, as well as the peculiar restriction of this tumor to humans.
Figure 1. Schematic view of the proposed viral etiology of prostate cancer.
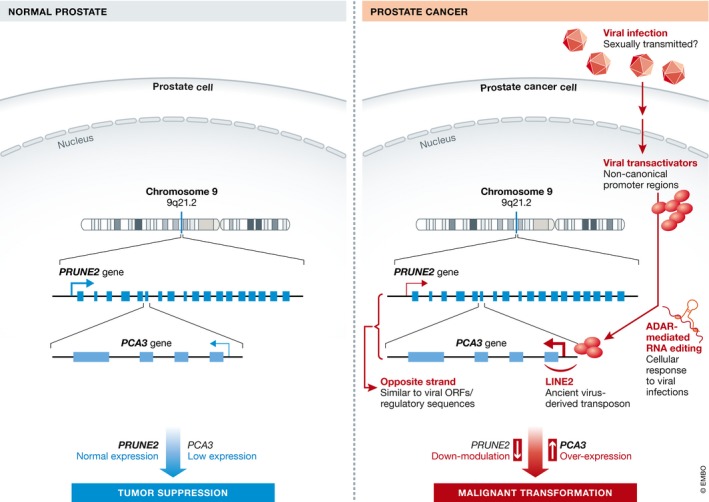
In the normal prostate, basal expression of PRUNE2 correlates with low PCA3 levels. In prostate cancer, a putative oncovirus could activate PCA3 expression through a direct transactivation of a cryptic, ancient virus‐derived promoter, resulting in downmodulation of PRUNE2, and leading to dysregulated cell proliferation and acquisition of malignant attributes.
Conflict of interest
RP and WA have a patent 15/500,686 licensed to Mbrace Therapeutics on the subject of the present manuscript. All other authors declare that they have no conflict of interest.
Acknowledgements
We thank Dr. Helen Pickersgill (Life Science Editors) for editorial assistance. The authors apologize to the many colleagues for not citing important work due to space limitations. AAT was supported by a fellowship from São Paulo Research Foundations (FAPESP, grant #2016/06069‐0). This work has been, in part, funded by a UNM Cancer Center Support Grant (P30 CA118100).
References
- Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N, Isaacs WB (1999) DD3: a new prostate‐specific gene, highly overexpressed in prostate cancer. Cancer Res 59: 5975–5979 [PubMed] [Google Scholar]
- Cao S, Moss W, O'Grady T, Concha M, Strong MJ, Wang X, Yu Y, Baddoo M, Zhang K, Fewell C et al (2015) New noncoding lytic transcripts derived from the Epstein‐Barr Virus latency origin of replication, oriP, are hyperedited, bind the paraspeckle protein, NONO/p54nrb, and support viral lytic transcription. J Virol 89: 7120–7132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D, Wojno K, Imperiale MJ (2008) BK virus as a cofactor in the etiology of prostate cancer in its early stages. J Virol 82: 2705–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marzo AM, Coffey DS, Nelson WG (1999) New concepts in tissue specificity for prostate cancer and benign prostatic hyperplasia. Urology 53: 29–42 [DOI] [PubMed] [Google Scholar]
- Dennis LK, Dawson DV (2002) Meta‐analysis of measures of sexual activity and prostate cancer. Epidemiology 13: 72–79 [DOI] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS (2008) Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319: 1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H (2002) Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2: 342–350 [DOI] [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C (2013) Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9: e1003470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Koonin EV (2017) Multiple origins of viral capsid proteins from cellular ancestors. Proc Natl Acad Sci USA 114: E2401–E2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X, Wang Y, Tian K, Ye F, Yin H, Zhao X, Xu H, Huang Y, Liu H, Hsieh JC et al (2017) Integrated host and viral transcriptome analyses reveal pathology and inflammatory response mechanisms to ALV‐J injection in SPF chickens. Sci Rep 7: 46156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Fu S, Sun LQ (2016) Viral noncoding RNAs in cancer biology. Adv Exp Med Biol 927: 367–389 [DOI] [PubMed] [Google Scholar]
- Moore PS, Chang Y (2014) The conundrum of causality in tumor virology: the cases of KSHV and MCV. Semin Cancer Biol 26: 4–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paprotka T, Delviks‐Frankenberry KA, Cingöz O, Martinez A, Kung HJ, Tepper CG, Hu WS, Fivash MJ Jr, Coffin JM, Pathak VK (2011) Recombinant origin of the retrovirus XMRV. Science 333: 97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis BS, Jungbluth AA, Frosina D, Holz M, Ritter E, Nakayama E, Ishida T, Obata Y, Carver B, Scher H et al (2013) Prostate cancer progression correlates with increased humoral immune response to a human endogenous retrovirus GAG protein. Clin Cancer Res 19: 6112–6125 [DOI] [PubMed] [Google Scholar]
- Salameh A, Lee AK, Cardo‐Vila M, Nunes DN, Efstathiou E, Staquicini FI, Dobroff AS, Marchiò S, Navone NM, Hosoya H et al (2015) PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad Sci USA 112: 8403–8408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalken JA, Hessels D, Verhaegh G (2003) New targets for therapy in prostate cancer: differential display code 3 (DD3(PCA3)), a highly prostate cancer‐specific gene. Urology 62: 34–43 [DOI] [PubMed] [Google Scholar]


