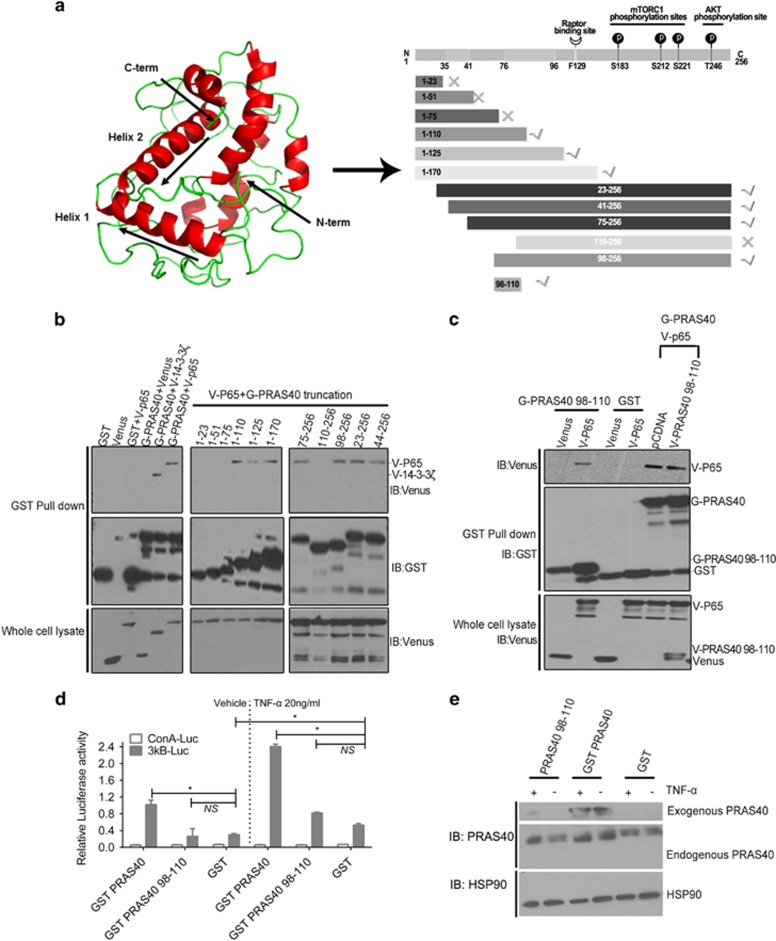
Figure 4.
PRAS40 98-110 is sufficient but not required for PRAS40–P65 association. (a) The PRAS40 structure was predicted by the I-TASSER algorithm and visualized using the PyMOL software (Left). The truncations of PRAS40 were produced based on the predicted structure of PRAS40 (Right). ✓ means could interact with P65; × means could not interact with P65 according to the results of a GST co-precipitation assay (b, c). (b) Truncations of PRAS40 with GST tag protein were co-transfected withVenus-P65 (V-P65, P65 protein with the Venus-Flag tag). The GST, Venus and truncations of PRAS40 (1-23, 1-51,1-75 and 110-256) could not co-precipitate P65 (lanes 1–3). However, the full length of PRAS40 and the truncations, including 1-110, 1-125, 1-170, 75-256, 98-256, 23-256 and 44-256 of PRAS40, could co-precipitate P65. 14-3-3ζwas used as the positive control for PRAS40 binding in this experiment. (c) The small truncation 98-110 of PRAS40 could co-precipitate P65 (lane 2 vs lane 1). But it could not significantly affect the co-precipitation of P65 by full-length PRAS40 (lane 6 vs lane 5). Lanes 3 and 4 were negative controls. (d) The 98-100 truncation of PRAS40 did not significantly regulate NF-κB transcriptional activity with or without tumor necrosis factor-α (TNF-α) stimulation (20 ng/ml, 2 h). (e) The proteins in the 3κB-Luc group cells were blotted to validate the transfection.