Figure 1.
Network Inference from Single-Cell Data
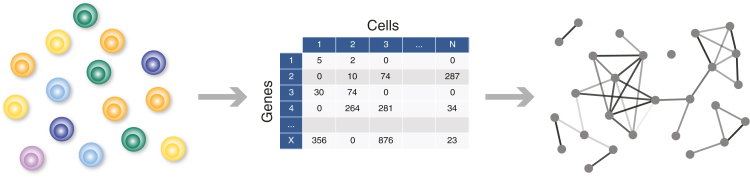
Single-cell transcriptomic data quantify mRNA species present inside individual cells. By considering pairs (or triplets, quadruplets, etc.) of mRNA species, we can test for statistical relationships among them. These dependencies may reflect coordinated gene expression of these pairs (or groups) of genes, resulting from gene regulatory interactions or co-regulation. Once such sets of genes that jointly change in expression are known, other statistical, bioinformatic, or text-mining analyses can be used to identify likely transcriptional regulators for these sets of genes. By iterating such in silico analyses with further, targeted experimental studies, we can, in principle, build up a representation of the gene regulatory network.