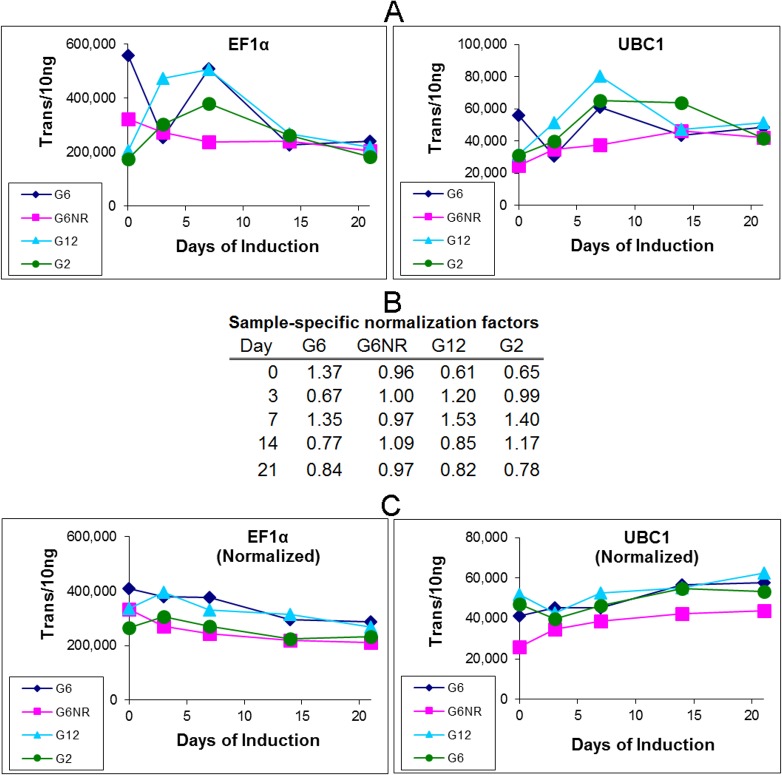
Fig 1. Intra-series normalization based on sample-specific differences in reference gene transcript quantity.
(A) Absolute transcripts quantities of two reference genes at five induction time points for each of the four explant types (G12 and G2: nonresponsive genotypes, G6 and G6NR: responsive and nonresponsive G6 trees, respectively). (B) Sample-specific normalization factors were generated by first converting reference gene transcript quantities within each individual sample into a fractional value relative to their respective sample series’ average, followed by averaging the two reference gene values. (C). Reference gene transcript quantities from A were divided by each respective sample-specific normalization factor from B to generate normalized quantities. Note that a central attribute of this approach is that normalization can be applied without loss of absolute scale.