Abstract
Telomeres are specialized chromatin structures that protect chromosome ends from dangerous processing events. In most tissues, telomeres shorten with each round of cell division, placing a finite limit on cell growth. In rapidly dividing cells, including the majority of human cancers, cells bypass this growth limit through telomerase-catalyzed maintenance of telomere length. The dynamic properties of telomeres and telomerase render them difficult to study using ensemble biochemical and structural techniques. This review describes single-molecule approaches to studying how individual components of telomeres and telomerase contribute to function. Single-molecule methods provide a window into the complex nature of telomeres and telomerase by permitting researchers to directly visualize and manipulate the individual protein, DNA, and RNA molecules required for telomere function. The work reviewed in this article highlights how single-molecule techniques have been utilized to investigate the function of telomeres and telomerase.
Keywords: telomerase, telomere, single-molecule biophysics
INTRODUCTION
Early in the evolution of single-celled and multicellular eukaryotic organisms, there was a transition away from circular genomes toward genomes composed of linear DNA molecules. Although this adaptation facilitated the development of modern eukaryotic genomes with increased complexity, it also introduced several interesting challenges for genomic stability and maintenance. First, cells harboring linear chromosomes must differentiate natural chromosome ends from bona fide sites of DNA damage. A second complication of the linear genome arises from the inability of the conventional replication machinery to complete the important task of copying the genetic material prior to cell division (82, 117). Thus, a cell with a linear genome must protect chromosome ends from unwanted recognition by DNA repair systems, as well as adapt to the incomplete duplication of the genetic material during replication. The ubiquitous solution to these two fundamental biological problems is the telomere, a specialized chromatin complex that safeguards the ends of eukaryotic linear genomes (68, 73).
The foundation of telomere structure is a highly repetitive DNA sequence. In human cells, telomeres consist of direct sequence repeats arranged into a long double-stranded region followed by a relatively short 3′ single-stranded guanine-rich (G-rich) tail (Figure 1a). These DNA sequences are unique to telomeric regions of the genome and serve to recruit a collection of telomere-specific proteins that regulate telomere structure and function. Interactions between telomere DNA and the multiprotein shelterin complex ensure chromosome ends evade unwanted recognition by DNA-damage response machinery (24). In addition, components of the shelterin complex are thought to modulate the functional state(s) and accessibility of telomeres to transacting factors (35). Although the precise mechanism of shelterin-mediated telomere protection is not completely understood, cells possessing dysfunctional shelterin complexes respond swiftly by halting cell division (senescence) or inducing programmed cell death (apoptosis) (40, 41, 120). This fact highlights the intimate relationship between telomere chromatin structure and the proliferative capacity of rapidly dividing cells. In a related manner, telomere length must also be maintained above a threshold limit to evade cell growth arrest (41). However, the inability of the conventional DNA replication machinery to completely copy the telomere 3′ end, often termed the end replication problem, causes a progressive shortening of telomere DNA with each round of cell division (82, 117). Consequently, telomere shortening effectively acts as a brake for rapidly proliferating cells, halting cellular growth when telomeres have reached a critically short length.
Figure 1.
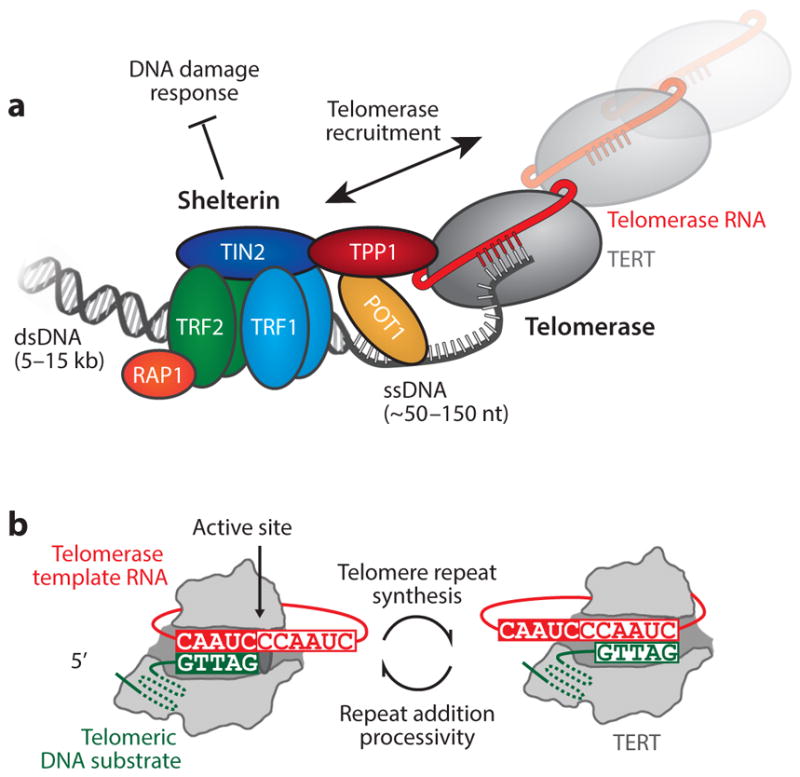
(a) Schematic diagram of shelterin and telomerase at the telomere. The shelterin complex binds to double-stranded telomeric DNA through TRF1 and TRF2 homodimers. TIN2 recruits the POT1–TPP1 heterodimer to the TRF complex. POT1 binds to the single-stranded, guanine-rich tail. These proteins prevent the DNA-damage response mechanisms from identifying telomeres as double-stranded breaks. An interaction between TPP1 and the N-terminal domain of telomerase regulates recruitment of telomerase to the telomeres. (b) The telomerase catalytic cycle. Telomerase catalysis begins with the 3′ end of the DNA substrate ( green) bound in the active site of the telomerase ribonucleoprotein (RNP) complex. The DNA primer aligns with the RNA template (red ) through Watson-Crick base pairing, providing the telomerase reverse transcriptase (TERT, gray) with an RNA–DNA hybrid substrate. Upon completion of a single telomere DNA repeat, the product–template hybrid must dissociate, realign, and reenter the TERT active site to support repeat addition processivity. Abbreviations: dsDNA, double-stranded DNA; ssDNA, single-stranded DNA.
Single-celled eukaryotic organisms, such as ciliated protozoans and yeasts, as well as rapidly dividing cell types within multicellular organisms, have found ways to circumvent the growth limitation imposed by the end replication problem. In the majority of cases, the solution is de novo telomere DNA synthesis by telomerase (38). Telomerase is a ribonucleoprotein (RNP) complex minimally composed of the telomerase reverse transcriptase (TERT) protein and a telomerase RNA (TR) (32, 75). To extend telomeres, telomerase binds to the 3′ single-stranded DNA (ssDNA) tail and uses a short segment within TR as a template for a TERT-catalyzed reverse transcription reaction (Figure 1b). Telomerase is unique among reverse transcriptases in its ability to catalyze multiple rounds of telomere DNA repeat synthesis prior to dissociating from its DNA substrate. The processive repeat addition activity of telomerase relies upon the functional interdependence of specialized protein and RNA domains, as well as unique structural properties of telomere DNA.
The direct link between telomere dysfunction and human disease has fueled intense interest toward understanding the complex interplay between the telomere, the telomerase enzyme, and the many additional cellular factors that contribute to telomere regulation. Exciting advances have been made in recent years in the area of telomere and telomerase structural biology [reviewed in this volume by Chan et al. (14a)], providing high-resolution structural data of telomere and telomerase subcomplexes, as well as electron microscopy (EM) reconstructions of the Tetrahymena thermophila and human telomerase holoenzymes (49, 67, 70, 94). At the same time, the single-molecule biophysics toolkit has become sufficiently established that many researchers have begun to develop assays that directly probe the dynamic and heterogeneous properties of telomeres and telomerase. Moreover, given the significant challenges that come with making telomerase enzymes for structural and biophysical studies, single-molecule techniques have found an important role to play in providing unique insight into the structural properties of telomerase interacting with telomere DNA substrates. In this review, we describe how single-molecule techniques have been employed to study telomeres and telomerase, with a focus on the novel insights that have been gained. This review does not attempt to describe each of the single-molecule methods in technical detail; therefore, we refer the reader to a number of excellent reviews on these subjects (25, 36, 72, 76, 92, 99, 109). Finally, we conclude with some future perspectives on how the continued use of single-molecule biophysical assays, together with improved structural data, may contribute to the ultimate goal of understanding the complexities of telomeres and telomerase.
TELOMERE DNA STRUCTURE AND DYNAMICS
The G-rich DNA repeat sequences (T2AG3 in humans) found at telomeres have a propensity to fold into unique secondary structures called G-quadruplexes (GQs) (Figure 2a). Human telomere DNA GQs are thought to play a role in regulating telomere homeostasis, and small-molecule ligands that selectively bind and stabilize telomere DNA GQs have shown promise as anticancer drugs (79, 89). Thus, intensive efforts have been made to better understand the structure and function of telomere DNA GQs. The first solution structure of a human telomere DNA GQ revealed a fundamental structural architecture in which guanine bases are hydrogen bonded in a planar quartet geometry and coordinate a single centrally located monovalent cation (116). Three adjacent intramolecular G-quartets can interact via stacking interactions and are topologically linked by short intervening DNA loop sequences (103). Analysis of telomere DNA GQs is complicated by the fact that a single GQ-forming sequence has the capacity to fold into multiple, nearly iso-energetic, structural variants that are distinguished by the topology of the DNA backbone (Figure 2a) (23). Thus, the study of telomere GQs represents an ideal application of single-molecule analysis, which eliminates ensemble averaging and thereby permits direct measurement of the folding energetics and kinetics of these heterogeneous and dynamic structures.
Figure 2.
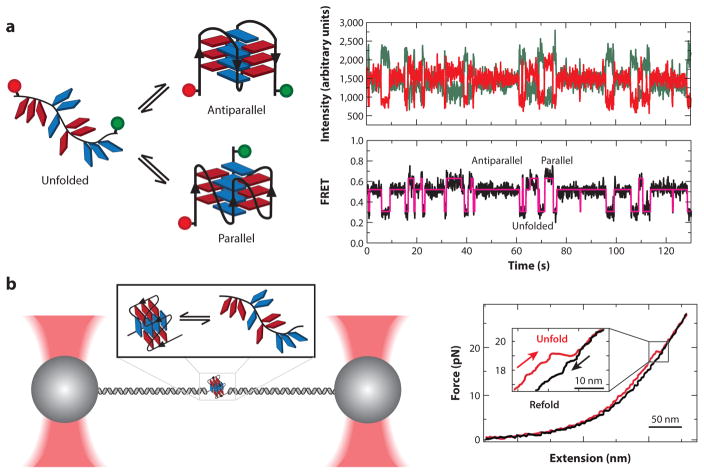
Single-molecule observation and mechanical manipulation of G-quadruplexes (GQs). (a) A doubly dye-labeled DNA construct is used in a single-molecule Förster resonance energy transfer (smFRET) experiment to measure the folding dynamics of a GQ. The smFRET signature reveals dynamic interconversion of the GQ between multiple structural states after passing through an unfolded intermediate. Panel a adapted with permission from Reference 63. (b) Optical tweezers are used to mechanically unfold a telomere GQ. Upon relaxation of the force, the single-stranded DNA can refold into the native GQ state. Analysis of the unfolding/refolding forces provides information about the energy landscape of folding. Panel b adapted with permission from Reference 26.
A wide variety of single-molecule techniques have been employed to investigate aspects of telomere DNA GQ folding. These techniques include single-molecule Förster resonance energy transfer (smFRET) (48, 57–59, 64, 74, 77, 80, 81, 110, 124), optical tweezers (26, 27, 54, 98), magnetic tweezers (125, 126), combined smFRET–magnetic tweezers (63), atomic force microscopy (AFM) (66), tethered particle motion assays (18, 62), and protein nanopores (4–6, 102, 119). Each approach comes with its own advantages and drawbacks; however, the most widely employed experiments to date have used smFRET or optical/magnetic tweezers. Using smFRET, the dynamic properties of human telomere DNA have been directly interrogated under a variety of conditions, with distinct FRET states corresponding to unfolded and folded GQ variants (Figure 2a). These studies demonstrate the powerful capacity of single-molecule biophysical measurements to monitor folding kinetics of dynamic biological structures under equilibrium conditions. In this type of experiment, the distributions of times spent in each of the observed FRET states provide important information about the underlying kinetics of the GQ folding process (109). Indeed, such dwell time analysis was performed for telomere DNA GQs and revealed several kinetically distinct, and previously unidentified, structural intermediates in the folding pathway (58). Dissecting the folding pathway of telomere GQ structure in this manner provides a conceptual framework for rational design of small molecules intended to alter the GQ folding equilibrium, as well as for studies aimed at understanding the molecular mechanisms of telomere GQ–resolving proteins (see below).
In the cell, the intrinsic telomere GQ folding equilibrium is altered by DNA-binding proteins and enzymes that remodel DNA structure by applying piconewton scale forces. To better understand how GQ structures respond to such mechanical perturbations, force spectroscopy methods, such as optical and magnetic tweezers, are often utilized. These micromanipulation techniques permit the application of precisely controlled tensions to telomere DNA GQ structures, providing details about the free-energy landscape for the GQ folding/unfolding reaction (76, 109). Mechanical forces of ~20 pN are required to rupture telomere GQ structure (Figure 2b) (26). This amount of force is comparable to forces generated by the molecular motors that mediate transcription and replication, giving one the sense of the mechanical challenges that GQs may introduce. Moreover, the ability to apply stretching forces to different parts of the GQ structure provides detailed information about the specific topological variant being studied. Interestingly, the use of simple transition state theory to analyze the effect of force on telomere-DNA-GQ–folding kinetics revealed the mechanical unfolding pathway of the telomere DNA GQ is characterized by a short distance (<1 nm) to the transition state for the unfolding reaction (63). This mechanical unfolding response reflects a critical contribution of long-range interactions to the global stability of the GQ fold and suggests that telomere-associated proteins need only disrupt a few base pairs to destabilize GQ structures. Thus, the use of applied force as a controlled experimental parameter has provided a new avenue for exploring the structural properties of telomere DNA GQs.
Early single-molecule experiments that investigated the structure and dynamics of a single telomere GQ fold provided unprecedented insight into these dynamic structures. More recently, single-molecule experiments have explored the manner in which deviations from canonical telomere DNA sequence influences GQ folding. For example, the impact of sequence mutations (57, 80), change in connecting loop size (87), and DNA damage (6, 33, 119) on the stability of telomere DNA GQ structures have all been measured using single-molecule methods. Collectively, these studies indicate that relatively minor sequence perturbations can exert significant destabilizing effects on telomere GQ folding. A natural segue from studying individual telomere GQ folds was to explore the more complex scenario of longer telomere DNAs. To this end, an optical tweezers assay was used to probe higher-order GQ structure and revealed that stacking of individual GQs can enhance the stability of neighboring GQ folds (98). In a separate study using smFRET, long ssDNA tails composed of many telomere DNA repeats were shown to be exceedingly dynamic (46), which is consistent with early AFM imaging studies of similar DNA substrates (115). This result suggests that GQ structures are dynamically formed and resolved on physiologically relevant timescales and may represent a mechanism of regulating the accessibility of the telomere to trans-acting factors.
Much of the basic work investigating telomere GQ structure has been motivated by interest in developing potent GQ-binding compounds for use as potential telomerase inhibitors or agents that disrupt normal telomere homeostasis in cancer cells (89). Several single-molecule studies have measured the properties of telomere GQ structures when bound to GQ-binding drugs. One study using smFRET surprisingly revealed that GQ structures are highly dynamic even when bound to certain GQ ligands (48). This result highlights the power of single-molecule experiments to reveal details of molecular interactions and suggests that alternative design strategies may be needed to achieve better GQ-binding drugs. In other single-molecule work, the use of optical tweezers, AFM, or tethered particle motion assays provided a sensitive and direct measure of the binding energy of various GQ ligands (34, 54, 62). Most GQ-binding ligands that have been studied to date are proposed to interact with GQ structure primarily through base-stacking interactions. As such, it has remained a major challenge to develop GQ-binding ligands with the high affinity and specificity required for therapeutic applications. Excitingly, a recent smFRET study reported the binding behavior of a novel class of small helical compounds that bind telomere GQ structure through a series of spatially specific backbone interactions, resulting in binding affinities on par with the best GQ-binding ligands to date (74).
Since the original discovery that short single-stranded telomere DNA model substrates can fold into a menagerie of GQ structures, this fold has captured the interest of hundreds of research groups, including many biophysicists. Nevertheless, debate continues as to whether the GQ structures that form so readily in vitro are in fact representative of telomere DNA structure in the cell. Recently, several studies have reported direct detection of GQ structures in cells (90), both at the telomere and elsewhere in the genome, refueling interest in telomere DNA GQ structure and dynamics. In the future, it will be of interest to exploit the power of single-molecule methods to explore the possible formation of GQ structure within duplex telomere DNA (97), rather than focusing entirely on the G-rich single-stranded telomere DNA tail. Furthermore, an important ongoing area of inquiry will be to utilize the experimental and conceptual framework established by these early single-molecule studies on naked telomere DNA GQs to study the interaction with proteins and enzymes that bind and remodel these captivating structures. Lastly, knowledge gained through the detailed analysis of telomere DNA GQs can also be extended to understand the function of GQ-forming sequences elsewhere in the genome—for instance, as regulatory elements within gene promoters.
TELOMERE DNA–BINDING PROTEINS AND ENZYMES
Mammalian telomeres are typically ~5–15 kb in length, composed mostly of duplex DNA that terminates with an ~150-nucleotide 3′ G-rich tail. The highly repetitive DNA sequences found at telomeres distinguish these regions from the rest of the genome and serve to recruit a battery of telomere-specific proteins and enzymes. Telomeric chromatin shares certain features with other genomic loci, such as the presence of nucleosomes; however, a six-protein complex called shelterin is a defining component of telomeres (Figure 1a) (14, 24, 104, 112). The shelterin complex is composed of three sequence-specific DNA-binding proteins: the protection of telomeres 1 (POT1) protein, which binds ssDNA, and the telomere repeat binding factors 1 and 2 (TRF1 and TRF2, respectively), which bind to duplex telomere DNA. Additional components of the shelterin complex include the TRF-interacting nuclear protein 2 (TIN2), the TIN2–POT1-binding protein (TPP1), and the repressor activator protein 1 (RAP1). TIN2 can simultaneously interact with the different DNA-binding subunits of shelterin to bridge the single-stranded and duplex DNA regions of the telomere. TPP1 is found primarily as a stable subunit of the heterodimeric POT1–TPP1 complex that can also make bridging interactions between the duplex region of the telomere and the ssDNA tail. Moreover, the POT1–TPP1 complex also directly interacts with the telomerase enzyme, regulating its recruitment to telomeres and its catalytic properties (100, 114). Thus, the core subunits of the shelterin complex have the capacity to form a diverse network of interactions that collectively regulate the functional state(s) of the telomere.
Several additional telomere-interacting complexes may be dynamically recruited to telomeres directly or through interactions with components of the shelterin complex. For example, the CST complex is a heterotrimeric protein complex that shares many features with replication protein A (RPA), which is the general eukaryotic ssDNA-binding complex (71). CST protects telomeres independently of shelterin, contributes to telomerase regulation, and mediates aspects of telomere DNA replication (17). Other trans-acting factors required for telomere replication or recombination include the RecQ-family DNA helicases BLM and WRN and the recombination protein RAD51 (21). More recently, the long noncoding telomeric repeat-containing RNA (TERRA) has been implicated in telomere maintenance (22), providing yet another mechanism for regulating the functional state of the telomere. Despite the myriad factors implicated in telomere regulation, models for telomere functional states are grossly oversimplified, often invoking open or closed conformations that have differing levels of accessibility to processing machinery (35). Indeed, the complexity and dynamics of the telomere interaction network make characterizing the molecular details of these telomere functional states challenging. This challenge makes single-molecule techniques, which are capable of dissecting the structure and dynamics of complex biological systems, a perfect match for analyzing telomere–protein interactions.
Among the earliest in singulo experiments performed on telomeres were the landmark EM experiments by Griffith et al. (39), which provided a glimpse of higher-order telomere structure. These studies revealed that TRF2 remodels linear telomere DNA fragments into large loops (T-loops) that are formed by the invasion of the 3′ ssDNA telomeric tail into the duplex region of the telomere, thus sequestering chromosome ends away from DNA-damage repair enzymes. The recent development of single-molecule fluorescence imaging techniques that permit researchers to achieve spatial resolutions well below the diffraction limit has opened up new avenues for studying telomere chromatin organization (44). For example, superresolution fluorescence imaging of telomeres demonstrated that conditional knockdown of TRF2, but not other shelterin subunits, disrupted the formation of T-loops, consistent with the earlier EM studies (28). In a separate study employing superresolution microscopy, the general level of telomere compaction was determined by measuring the volume occupied by individual telomeres within intact cells (7). In the future, the ability to monitor the structural state and dynamic protein occupancy of intact telomeres in living cells will undoubtedly provide new insights into telomere structure and function.
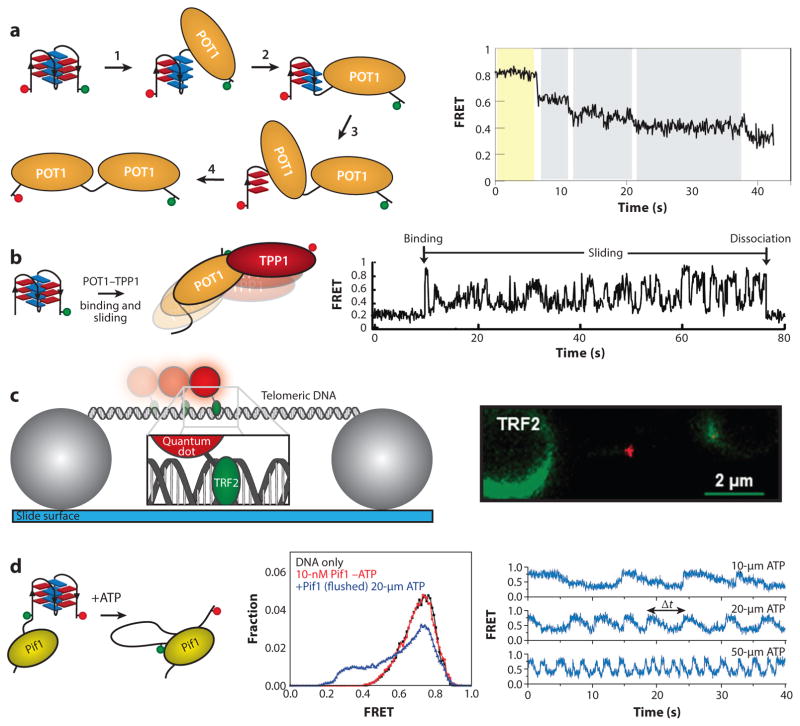
Single-molecule techniques have been used to directly analyze protein-binding dynamics on single-stranded telomere DNA substrates to understand the functional remodeling of the telomeres. For example, the binding of POT1 and the POT1–TPP1 heterodimer to ssDNA telomere substrates was monitored by smFRET (Figure 3a,b) (45). This work demonstrated that POT1 binds and sequentially unfolds a telomere DNA GQ structure through interaction with each of its individual OB (oligonucleotide–oligosaccharide) binding domains. In contrast, the POT1–TPP1 complex dynamically slides back and forth along telomere DNA substrates (Figure 3b). This sliding activity is similar to that reported for other OB-fold proteins, such as the single-stranded DNA-binding protein of Escherichia coli (93), and may underlie the enhanced processivity of telomerase when bound to the POT1–TPP1 complex (114). In a separate study, smFRET was used to show that telomere overhang length regulates the accessibility of telomere-associated proteins RAD51, WRN, and BLM (46). Interestingly, telomeric DNA substrates that had multiples-of-four repeats showed limited protein binding, whereas those with an odd number of repeats were more readily bound. In contrast, binding of the shelterin protein POT1 was independent of the number of telomere DNA repeats present. These experiments offer new insight into how the intrinsic structural properties of telomere DNA can regulate the binding of certain telomere-associated factors. In a related study, researchers again used smFRET to address the question of how single-stranded telomere DNA substrates are preferentially bound by the shelterin protein POT1 rather than the DNA-damage response protein RPA (86). The level of RPA in the cell is much higher than POT1, and RPA has been shown to readily bind and resolve a variety of GQ DNA structures (85, 87). However, it was shown that under physiological conditions, the POT1–TPP1 complex binds and protects telomere DNA GQs from RPA interaction by two orders of magnitude. These single-molecule experiments highlight the dynamic nature of telomere-DNA–protein interaction; yet, there is still much to be done. For example, single-molecule experiments recently showed that the CST complex can also resolve telomere GQ structure (65). Thus, it will be interesting to see how the CST complex influences the binding properties of POT1–TPP1 and/or RPA. Moreover, the use of multicolor single-molecule imaging will play an important role in dissecting the ssDNA-binding dynamics of complex mixtures of telomere-associated proteins.
Figure 3.
Telomere-associated proteins interact dynamically with telomere DNA. (a) A single-molecule Förster resonance energy transfer (smFRET) assay is used to probe the state of the telomere DNA quadruplex in the presence of POT1. The stepwise unfolding of a quadruplex by POT1 is observed as a series of consecutive decreases in FRET throughout the single-molecule trace. (b) Using the same experimental setup as in panel a, the heterodimeric POT1–TPP1 complex displays a dynamic sliding behavior once bound to the DNA substrate. Panels a and b adapted with permission from Reference 45. (c) A double-stranded telomere DNA tightrope is stretched between two immobilized beads. TRF2 protein conjugated to a quantum dot is added to solution, and single-molecule observation of dynamic TRF2 association with telomere DNA is made in real time. Comparison of telomere versus non-telomere DNA provides kinetic parameters about TRF2 diffusion along telomere DNA. Panel c adapted with permission from Reference 60. (d ) A schematic depiction of an smFRET assay for measuring quadruplex unfolding by the helicase Pif1. Unwinding of the quadruplex by Pif1 is ATP dependent and reversible. The sawtooth pattern represents binding by Pif1, followed by the repetitive unfolding of the quadruplex by the DNA reeling action of a single Pif1 enzyme. Panel d adapted with permission from Reference 127.
Single-molecule imaging has also provided new and interesting insight into the dynamics of the duplex DNA–binding proteins, TRF1 and TRF2. Using a novel DNA tightrope assay, researchers have directly monitored the binding and diffusion of fluorescently labeled TRF1 and TRF2 proteins on long duplex telomere DNA substrates (Figure 3c) (60). Unlike the static images of preformed TRF1– or TRF2–DNA complexes provided by EM and superresolution microscopy, these experiments provide a direct view of how these essential telomere-binding proteins find their sites of action. Interestingly, TRF proteins not only employ a one-dimensional diffusive search to find telomere DNA but continue to dynamically slide when bound to telomere DNA sequence. The dynamic binding behavior of the TRF proteins may underlie the assembly of diverse shelterin complexes, as well as interactions between the TRF proteins and non-shelterin proteins. Consistent with this notion, a follow-up study using the same imaging assay demonstrated that dynamic interactions between TRF1 and the chromosome cohesion subunit SA1 promote DNA–DNA pairing at sister telomeres (61). Interestingly, researchers recently used a similar DNA curtain assay to monitor in real time the binding properties of reconstituted mammalian shelterin complexes, concluding that shelterin localizes to telomere DNA primarily through a three-dimensional search process (31). In a series of separate imaging studies, AFM was used to demonstrate that TRF2, but not TRF1, wraps and condenses telomere DNA (3, 9, 51). TRF2 wrapping of DNA was shown to be chiral and therefore introduces compensatory superhelical strain into telomere DNA that was proposed to regulate DNA transactions at telomeres, such as strand invasion. Future single-molecule experiments aimed at measuring the dynamics of TRF2 wrapping and the energetics of strand invasion at telomeres will likely expand our appreciation of the dynamic properties of shelterin and its associated proteins.
In addition to proper function of shelterin proteins, telomere maintenance also requires the coordination of multiple enzymatic activities. For example, conventional replication machinery is responsible for copying most of the telomere during semiconservative replication. The highly repetitive property of telomere DNA sequences, together with the shelterin-mediated organization of the telomere (i.e., T-loops), introduces obstacles to the replication process (112). In fact, telomeres have been shown to resemble fragile sites during replication under certain conditions, requiring the activity of specific factors to help promote progression of the replication fork (101). An imaging technique called single-molecule analysis of replicated DNA (SMARD) permits mapping of replication fork stalling under various conditions. Using the SMARD technique, researchers demonstrated that the shelterin subunit TRF1 and two telomere-associated DNA helicases (BLM and RTEL1) are required to prevent fork stalling (29). In several separate studies, smFRET experiments were used to dissect the catalytic properties of the Pif1 and BLM helicases (127). Remarkably, both of these motor proteins display a so-called DNA patrolling activity on telomere DNA GQ substrates, wherein the helicase reiteratively unfolds the GQ structure by reeling in the 3′ tail and then releasing it to refold and be unwound again (Figure 3d). The periodic unwinding activity of GQ-resolving helicases discovered using single-molecule methods suggests a mechanism by which cellular enzymatic activities may effectively reduce the stability of GQ structures to facilitate maintenance of telomeres. Of course, the enzymatic activity most often associated with telomere maintenance is that of the reverse transcriptase enzyme telomerase (described in detail below). Recently, live-cell single-molecule imaging has been used to investigate how telomerase finds telomere DNA ends during the S-phase of the cell cycle (96). Direct telomerase labeling techniques permitted researchers to track telomerase complexes in real time, revealing that telomerase primarily employs a three-dimensional diffusive search to engage the telomere for extension. Moreover, the ability of telomerase to stably associate with telomere substrates was dependent on the shelterin protein TPP1, consistent with previous reports that demonstrated that direct TPP1–telomerase interactions mediate telomerase recruitment to telomeres (100).
Telomeres are dynamic nucleoprotein structures, from the unique structural properties of the G-rich telomere DNA repeats to the complex orchestration of all the proteins (and now TERRA RNA) required for telomere maintenance. Single-molecule methods have begun to shed new light on the structure and dynamics of telomeres; yet, there is much that remains to be done. For example, micromanipulation techniques may be used to better understand how shelterin proteins cooperate to remodel telomere structure and facilitate transactions at long duplex telomere DNA substrates. Another particularly rich area of inquiry will be the physical basis for the challenges imposed by repetitive telomere DNA sequences during replication. Although single-molecule analysis has provided a tremendous amount of insight into the mechanism of the bacterial replisome (111), methods for reconstituting the eukaryotic replisome for biophysical experiments are just beginning to emerge (78). With these powerful biochemical replication systems in place, it is inevitable that single-molecule approaches will contribute to the ultimate goal of understanding the detailed molecular mechanisms of telomere DNA replication and chromatin remodeling.
TELOMERASE STRUCTURE AND DYNAMICS
Rapidly dividing cells must combat the gradual telomere attrition that occurs during multiple rounds of cell division. In most cases, this is accomplished by telomerase-catalyzed addition of short sequence repeats onto the 3′ ssDNA tail of the telomere (43, 95). Telomerase is commonly upregulated in human cancers (53); thus, a detailed understanding of telomerase assembly and regulation holds direct biomedical significance. Moreover, telomerase is a member of a class of RNP enzymes that mediate a wide variety of cellular processes, ranging from protein synthesis on the ribosome to messenger RNA processing by the spliceosome. Each of these essential RNP complexes relies upon a precise interdependence of protein and RNA subunits; therefore, telomerase also serves as a powerful model system for understanding general principles that govern RNP structure and function. The telomerase catalytic cycle resembles a canonical reverse transcription mechanism of RNA-directed DNA synthesis; however, a catalytic substep unique to telomerase facilitates addition of multiple telomere DNA repeats during a single DNA-binding event (37). This activity is termed repeat addition processivity (RAP) and requires a series of coordinated conformational rearrangements to facilitate reiterative utilization of the integral TR template by the reverse transcriptase protein (Figure 1b). Conserved motifs in both the RNA and protein components of telomerase have been implicated in RAP (11, 30, 50, 55, 56, 91, 122), but the precise structural dynamics associated with this unique substep are still poorly understood. Furthermore, telomerase structure–function studies are complicated by challenges associated with generating sufficient quantities of homogeneous enzyme preparations required for traditional structural techniques. Thus, single-molecule techniques are particularly useful for dissecting the structure and dynamics that underlie telomerase function.
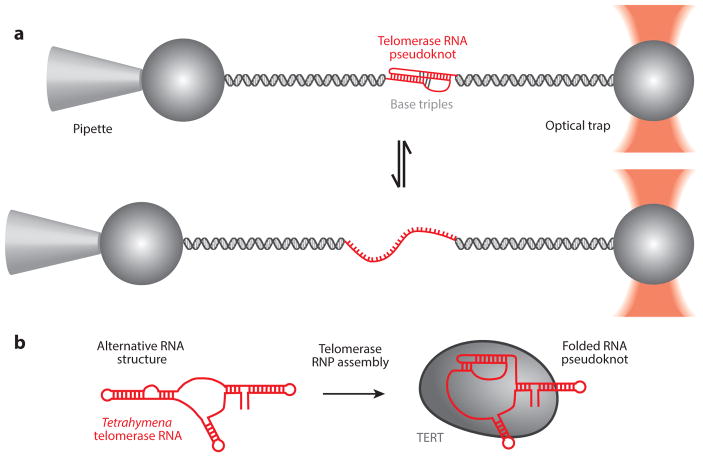
Telomerase RNAs not only provide the template for the reverse transcription reaction but also share several additional conserved structural motifs required for proper RNP assembly and function (107). For example, all telomerase RNAs possess a pseudoknot fold positioned 3′ of the RNA template. Mutations in the pseudoknot alter telomerase activity (12, 52, 106) and are linked to several genetically inherited human disorders characterized by symptoms of premature aging (113). Early structural studies of the human TR pseudoknot utilized a minimized construct amenable to high-resolution structure determination by nuclear magnetic resonance (NMR) (52, 108). The folding properties of this minimal RNA pseudoknot were subsequently analyzed at the single-molecule level using optical tweezers (15), providing a powerful model system for understanding how the complex RNA folding landscape must be manipulated during RNP assembly (Figure 4a). In an effort to study the folding properties of the full-length RNA pseudoknot, smFRET was used to show that physiological levels of magnesium promote human pseudoknot folding in the absence of the telomerase protein component (42). Interestingly, a separate study used smFRET to demonstrate that the T. thermophila pseudoknot motif is misfolded and requires protein binding to stabilize the native fold (Figure 4b) (69), consistent with biochemical and structural studies of the same system that characterized an alternative fold for this region of the RNA in the absence of the TERT protein (13, 19). Taken together, these results suggest that the larger vertebrate pseudoknot domain has evolved to fold independently during telomerase assembly. Once assembled into the telomerase RNP complex, the RNA pseudoknot was previously proposed to act as a molecular switch and undergo conformational rearrangements during processive telomerase activity (20). However, recent smFRET studies of the assembled telomerase RNP have shown that, in both humans and T. thermophila, the RNA pseudoknot appears stably folded throughout the catalytic cycle at the timescales probed in the experiments (69, 83). Although conformational changes at faster timescales cannot be excluded, these results are consistent with earlier biochemical analysis of the human RNA pseudoknot that supports a static pseudoknot structure (16) and leave open the important question of why this interesting RNA fold has been evolutionarily conserved.
Figure 4.
Folding dynamics of the telomerase RNA (TR) pseudoknot. (a) Using optical tweezers, the human telomerase pseudoknot was stretched to induce unfolding of the pseudoknot through base-pair shearing. Refolding of the pseudoknot was observed as the force is relaxed. Panel a adapted with permission from Reference 15. (b) Tetrahymena telomerase RNA pseudoknot folding during assembly into an active ribonucleoprotein (RNP) complex was monitored by single-molecule Förster resonance energy transfer (smFRET) (69). This result is consistent with models for an alternative RNA structure in the absence of telomerase proteins derived from biochemical and structural analyses (13, 19). Abbreviation: TERT, telomerase reverse transcriptase.
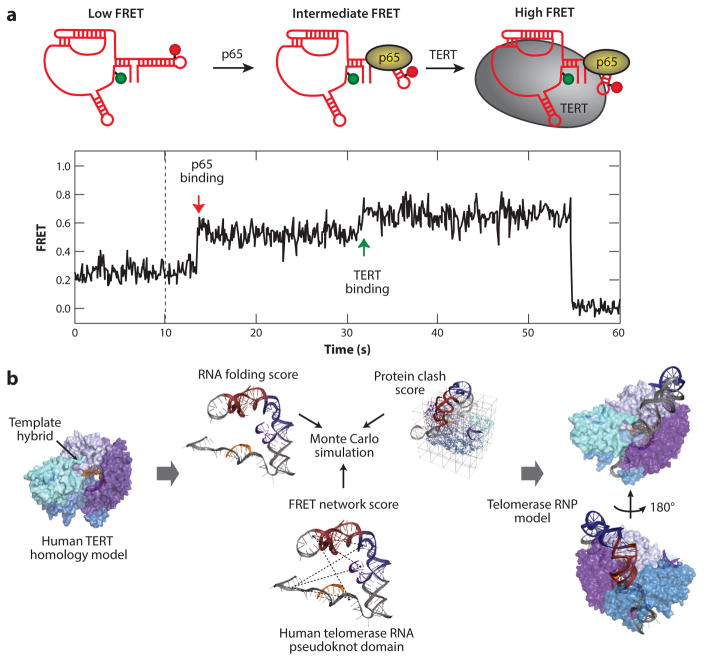
Single-molecule analysis has proven to be a powerful tool for visualizing individual telomerase complexes, providing the discrete stoichiometry of RNP components as well as a real-time visualization of RNP structure and assembly. Early studies of human telomerase used two-color coincidence detection to show that catalytically active telomerase functions as a monomer (2). Subsequent to this work, a low-resolution EM reconstruction and accompanying biochemical analysis of the human telomerase holoenzyme suggested the complex functions as an obligate dimer (94), resurrecting much earlier models for telomerase dimerization (8, 118). In a more recent study, rigorous single-molecule colocalization studies compared the various telomerase reconstitution and purification protocols reported in the literature (123). The results of this work confirmed telomerase is catalytically active as a monomer and also mapped a region within the telomerase reverse transcriptase protein subunit that is responsible for the propensity of purified telomerase to aggregate into higher-order structures. Several additional single-molecule studies have focused on using in vitro reconstituted monomeric telomerase complexes to study RNP assembly and dynamics. For example, smFRET was used to monitor protein-mediated RNA folding during the assembly pathway of T. thermophila telomerase (Figure 5a) (105). This work revealed that binding of the telomerase holoenzyme protein p65 induces a bend in an essential RNA stem-loop, which in turn promotes the hierarchical assembly of the telomerase reverse transcriptase subunit. More recently, a comprehensive set of smFRET measurements involving human TR were acquired within the catalytically active human telomerase RNP. These smFRET measurements provided distance constraints that guided Rosetta modeling of the overall architecture and dynamics of human TR during catalysis (Figure 5b) (83). This work mapped the location of the pseudoknot fold within the RNP complex and revealed large-scale conformational rearrangements of this RNA domain during catalysis, hinting at a potential function of the pseudoknot as a regulator of protein conformation through physical connectivity to the template element.
Figure 5.
Assembly of the telomerase ribonucleoprotein (RNP) visualized by single-molecule Förster resonance energy transfer (smFRET). (a) In Tetrahymena thermophila telomerase, an smFRET pair was used to measure conformational changes in telomerase RNA (TR) during RNP assembly. Sequential binding of p65 and then telomerase reverse transcriptase (TERT) induces stepwise folding of TR visualized by consecutive increases in the FRET signal. Panel a adapted with permission from Reference 105. (b) A schematic pipeline for molecular modeling of the human telomerase complex. A homology model was generated for human TERT and used as a rigid modeling scaffold. smFRET distance constraints were used to guide Rosetta modeling of the RNA pseudoknot domain within the RNP. Convergent models of the RNP complex were produced, providing insight into human telomerase RNP architecture. Panel b adapted with permission from Reference 83.
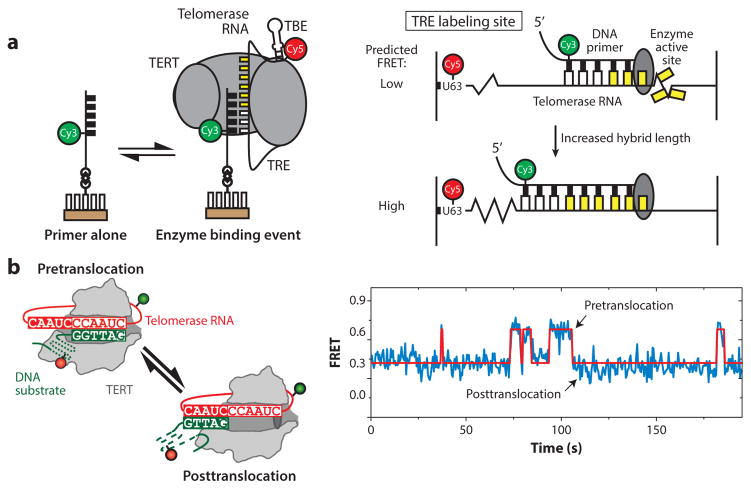
The complex steps required for each round of the telomerase catalytic cycle imply a high degree of conformational flexibility within the enzyme. Characterizing these internal structural dynamics during telomerase catalysis has been an exciting area for the application of single-molecule methods. For example, a series of smFRET experiments on Tetrahymena telomerase revealed that extension of the template hybrid induces motion of the template through the active site until stretching of the RNA is no longer possible because of an upstream protein–RNA anchor point, thereby serving to define the template boundary (Figure 6a) (10). Another long-standing question in the telomerase field has been how the nascent template–product hybrid dissociates to permit realignment with the downstream region of the RNA template. Recently, smFRET was used to monitor DNA-binding dynamics within human telomerase at various stages of telomere repeat synthesis, revealing internal structural dynamics between the 3′ end of the DNA and the RNA template (Figure 6b) (84). This result implies that telomerase structural changes triggered by completion of a telomere repeat facilitate realignment of the DNA primer to begin the next catalytic cycle. In a related follow-up study using the T. thermophila system, the conserved telomerase essential N-terminal domain of TERT was shown to be essential for stabilization of the short nascent hybrid created following realignment of the DNA product (1), highlighting the coordinated roles of protein, RNA, and DNA in supporting processive telomere repeat addition.
Figure 6.
Intramolecular telomerase dynamics. (a) Single-molecule Förster resonance energy transfer (smFRET) assay measuring internal movement of telomerase RNA throughout the catalytic cycle. RNA sequences flanking the template exhibit reciprocal compression and expansion motion during telomere DNA synthesis. Panel a adapted with permission from Reference 10. (b) DNA substrate dynamics in human telomerase observed by smFRET. Upon completion of one round of telomere repeat synthesis, the 3′ end of the DNA primer exhibits rapid dynamics between the two possible base-pairing registers with the RNA template. Kinetic analysis of these single-molecule data suggests that template hybrid realignment is much faster than the rate-limiting step of telomerase translocation required during repeat addition processivity. Panel b adapted with permission from Reference 84. Abbreviations: TBE, template boundary element; TERT, telomerase reverse transcriptase; TRE, telomeric repeat elongation.
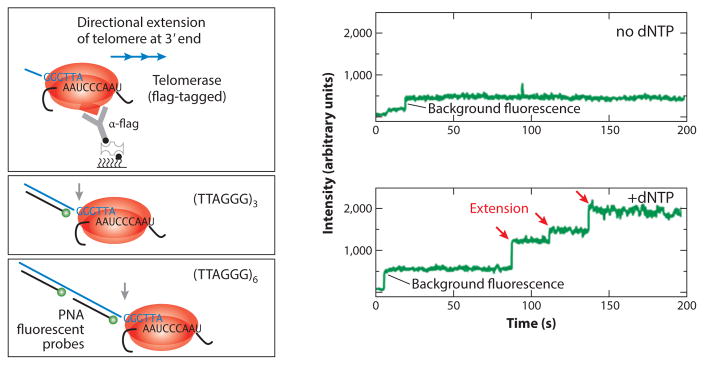
An ultimate goal of telomerase biophysical analysis is to directly correlate structural dynamics with specific stages of catalysis. Toward this end, researchers have begun to explore ways of using single-molecule methods to directly monitor telomerase catalysis. One early incarnation of this approach employed two-color coincidence detection to detect telomerase activity at the single-molecule level (88). In a separate smFRET study, products of individual telomerase–substrate-binding complexes were probed using a short complementary detection oligo, and the dissociation kinetics were used to decipher the length of the DNA products (121). However, both of these early approaches suffered from the inability to report on telomerase catalysis in real time. More recently, simultaneous imaging of telomerase–DNA complexes in the presence of dye-labeled peptide nucleic acid probes directly revealed the kinetic properties of individual telomerase enzymes from mammalian cells (Figure 7) (47). Variations of this real-time assay will likely be integrated with other structure-probing measurements at the single-molecule level to directly characterize how internal structural dynamics contribute to telomerase catalysis.
Figure 7.
Real-time telomerase activity assay at the single-molecule level. Human telomerase immobilized on a surface can bind to and extend a telomere DNA primer. As telomerase extends the primer processively, the telomere DNA repeats are extruded out of the telomerase complex and bound by a fluorescently labeled peptide nucleic acid (PNA) probe. Consecutive binding of multiple PNA probes generates a stepping pattern in the single-molecule fluorescence trace, providing real-time kinetic parameters governing the telomerase catalytic process. Adapted with permission from Reference 47. Abbreviation: dNTP, deoxynucleotide.
FUTURE PERSPECTIVES
The power of single-molecule science lies in the ability to resolve dynamic and heterogeneous properties of complex biological systems. In this review, we have highlighted the ways in which single-molecule methods can complement other biochemical and structural approaches to provide unique insight into molecular mechanisms that underlie telomere and telomerase function. In the future, continued advancement of single-molecule methods will enable researchers to probe the structural polymorphism of telomeres and telomerase in living cells. Detailed biophysical analyses of different shelterin subcomplexes will inform models for how these telomere-associated proteins regulate the functional state(s) of telomeres. In addition, the use of single-molecule techniques will undoubtedly contribute to studies of how the direct repeat sequences at telomeres impose challenges to DNA replication machinery. With respect to telomerase, improved structural models emerging from cryoEM, NMR, and crystallography studies will usher in a new era of rationally designed single-molecule experiments to dissect the precise structural rearrangements required for telomerase catalysis. Finally, new detection schemes for real-time telomerase activity detection, coupled with single-molecule structure-probing methods, will provide the opportunity to directly relate telomerase structural dynamics with specific kinetic substeps of the catalytic cycle.
Acknowledgments
We apologize to any authors whose work we were unable to include because of space constraints. M.D.S. is supported by NIH grant RO1GM095850; J.W.P. is supported by NSF grant DGE 0809125.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Akiyama BM, Parks JW, Stone MD. The telomerase essential N-terminal domain promotes DNA synthesis by stabilizing short RNA–DNA hybrids. Nucleic Acids Res. 2015;43:5537–49. doi: 10.1093/nar/gkv406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alves D, Li H, Codrington R, Orte A, Ren X, et al. Single-molecule analysis of human telomerase monomer. Nat Chem Biol. 2008;4:287–89. doi: 10.1038/nchembio.82. [DOI] [PubMed] [Google Scholar]
- 3.Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, et al. A topological mechanism for TRF2-enhanced strand invasion. Nat Struct Mol Biol. 2007;14:147–54. doi: 10.1038/nsmb1192. [DOI] [PubMed] [Google Scholar]
- 4.An N, Fleming AM, Burrows CJ. Interactions of the human telomere sequence with the nanocavity of the α-hemolysin ion channel reveal structure-dependent electrical signatures for hybrid folds. J Am Chem Soc. 2013;135:8562–70. doi: 10.1021/ja400973m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.An N, Fleming AM, Middleton EG, Burrows CJ. Single-molecule investigation of G-quadruplex folds of the human telomere sequence in a protein nanocavity. PNAS. 2014;111:14325–31. doi: 10.1073/pnas.1415944111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An N, Fleming AM, White HS, Burrows CJ. Nanopore detection of 8-oxoguanine in the human telomere repeat sequence. ACS Nano. 2015;9:4296–307. doi: 10.1021/acsnano.5b00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bandaria JN, Qin P, Berk V, Chu S, Yildiz A. Shelterin protects chromosome ends by compacting telomeric chromatin. Cell. 2016;164:735–46. doi: 10.1016/j.cell.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beattie TL, Zhou W, Robinson MO, Harrington L. Functional multimerization of the human telomerase reverse transcriptase. Mol Cell Biol. 2001;21:6151–60. doi: 10.1128/MCB.21.18.6151-6160.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benarroch-Popivker D, Pisano S, Mendez-Bermudez A, Lototska L, Kaur P, et al. TRF2-mediated control of telomere DNA topology as a mechanism for chromosome-end protection. Mol Cell. 2016;61:274–86. doi: 10.1016/j.molcel.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman AJ, Akiyama BM, Stone MD, Cech TR. The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis. Nat Struct Mol Biol. 2011;18:1371–75. doi: 10.1038/nsmb.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryan TM, Goodrich KJ, Cech TR. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol Cell. 2000;6:493–99. doi: 10.1016/s1097-2765(00)00048-4. [DOI] [PubMed] [Google Scholar]
- 12.Cash DD, Cohen-Zontag O, Kim N-K, Shefer K, Brown Y, et al. Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo. PNAS. 2013;110:10970–75. doi: 10.1073/pnas.1309590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cash DD, Feigon J. Structure and folding of the Tetrahymena telomerase RNA pseudoknot. Nucleic Acids Res. 2017;45:482–95. doi: 10.1093/nar/gkw1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–79. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 14a.Chan H, Wang Y, Feigon J. Progress in human and Tetrahymena telomerase structure determination. Annu Rev Biophys. 2017;46:199–225. doi: 10.1146/annurev-biophys-062215-011140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen G, Wen J-D, Tinoco I., Jr Single-molecule mechanical unfolding and folding of a pseudoknot in human telomerase RNA. RNA. 2007;13:2175–88. doi: 10.1261/rna.676707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen JL, Greider CW. Functional analysis of the pseudoknot structure in human telomerase RNA. PNAS. 2005;102:8080–85. doi: 10.1073/pnas.0502259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L-Y, Redon S, Lingner J. The human CST complex is a terminator of telomerase activity. Nature. 2012;488:540–44. doi: 10.1038/nature11269. [DOI] [PubMed] [Google Scholar]
- 18.Chu J-F, Chang T-C, Li H-W. Single-molecule TPM studies on the conversion of human telomeric DNA. Biophys J. 2010;98:1608–16. doi: 10.1016/j.bpj.2009.12.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole DI, Legassie JD, Bonifacio LN, Sekaran VG, Ding F, et al. New models of Tetrahymena telomerase RNA from experimentally derived constraints and modeling. J Am Chem Soc. 2012;134:20070–80. doi: 10.1021/ja305636u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comolli LR, Smirnov I, Xu L, Blackburn EH, James TL. A molecular switch underlies a human telomerase disease. PNAS. 2002;99:16998–7003. doi: 10.1073/pnas.262663599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem. 2014;83:519–52. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cusanelli E, Chartrand P. Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front Genet. 2015;6:143. doi: 10.3389/fgene.2015.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai J, Carver M, Yang D. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90:1172–83. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–10. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 25.Deniz AA, Mukhopadhyay S, Lemke EA. Single-molecule biophysics: at the interface of biology, physics and chemistry. J R Soc Interface. 2008;5:15–45. doi: 10.1098/rsif.2007.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhakal S, Cui Y, Koirala D, Ghimire C, Kushwaha S, et al. Structural and mechanical properties of individual human telomeric G-quadruplexes in molecularly crowded solutions. Nucleic Acids Res. 2013;41:3915–23. doi: 10.1093/nar/gkt038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhakal S, Schonhoft JD, Koirala D, Yu Z, Basu S, Mao H. Coexistence of an ILPR i-motif and a partially folded structure with comparable mechanical stability revealed at the single-molecule level. J Am Chem Soc. 2010;132:8991–97. doi: 10.1021/ja100944j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155:345–56. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drosopoulos WC, Kosiyatrakul ST, Schildkraut CL. BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J Cell Biol. 2015;210:191–208. doi: 10.1083/jcb.201410061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckert B, Collins K. Roles of telomerase reverse transcriptase N-terminal domain in assembly and activity of Tetrahymena telomerase holoenzyme. J Biol Chem. 2012;287:12805–14. doi: 10.1074/jbc.M112.339853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erdel F, Kratz K, Willcox S, Griffith JD, Greene EC, de Lange T. Telomere recognition and assembly mechanism of mammalian shelterin. Cell Rep. 2017;18:41–53. doi: 10.1016/j.celrep.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, et al. The RNA component of human telomerase. Science. 1995;269:1236–41. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 33.Fouquerel E, Lormand J, Bose A, Lee H-T, Kim GS, et al. Oxidative guanine base damage regulates human telomerase activity. Nat Struct Mol Biol. 2016;23:1092–100. doi: 10.1038/nsmb.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Funayama R, Nakahara Y, Kado S, Tanaka M, Kimura K. A single-molecule force-spectroscopic study on stabilization of G-quadruplex DNA by a telomerase inhibitor. Analyst. 2014;139:4037–43. doi: 10.1039/c4an00439f. [DOI] [PubMed] [Google Scholar]
- 35.Galati A, Micheli E, Cacchione S. Chromatin structure in telomere dynamics. Front Oncol. 2013;3:46. doi: 10.3389/fonc.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenleaf WJ, Woodside MT, Block SM. High-resolution, single-molecule measurements of biomolecular motion. Annu Rev Biophys Biomol Struct. 2007;36:171–90. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greider CW. Telomerase is processive. Mol Cell Biol. 1991;11:4572–80. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–13. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 39.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–14. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 40.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–36. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 41.Hayflick L. The cell biology of aging. Clin Geriatr Med. 1985;1:15–27. [PubMed] [Google Scholar]
- 42.Hengesbach M, Kim N-K, Feigon J, Stone MD. Single-molecule FRET reveals the folding dynamics of the human telomerase RNA pseudoknot domain. Angew Chem Int Ed Engl. 2012;51:5876–79. doi: 10.1002/anie.201200526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hockemeyer D, Collins K. Control of telomerase action at human telomeres. Nat Struct Mol Biol. 2015;22:848–52. doi: 10.1038/nsmb.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010;143:1047–58. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hwang H, Buncher N, Opresko PL, Myong S. POT1-TPP1 regulates telomeric overhang structural dynamics. Structure. 2012;20:1872–80. doi: 10.1016/j.str.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang H, Kreig A, Calvert J, Lormand J, Kwon Y, et al. Telomeric overhang length determines structural dynamics and accessibility to telomerase and ALT-associated proteins. Structure. 2014;22:842–53. doi: 10.1016/j.str.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hwang H, Opresko P, Myong S. Single-molecule real-time detection of telomerase extension activity. Sci Rep. 2014;4:6391. doi: 10.1038/srep06391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jena PV, Shirude PS, Okumus B, Laxmi-Reddy K, Godde F, et al. G-quadruplex DNA bound by a synthetic ligand is highly dynamic. J Am Chem Soc. 2009;131:12522–23. doi: 10.1021/ja903408r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang J, Chan H, Cash DD, Miracco EJ, Ogorzalek Loo RR, et al. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science. 2015;350:aab4070. doi: 10.1126/science.aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jurczyluk J, Nouwens AS, Holien JK, Adams TE, Lovrecz GO, et al. Direct involvement of the TEN domain at the active site of human telomerase. Nucleic Acids Res. 2011;39:1774–88. doi: 10.1093/nar/gkq1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaur P, Wu D, Lin J, Countryman P, Bradford KC, et al. Enhanced electrostatic force microscopy reveals higher-order DNA looping mediated by the telomeric protein TRF2. Sci Rep. 2016;6:20513. doi: 10.1038/srep20513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim N-K, Zhang Q, Zhou J, Theimer CA, Peterson RD, Feigon J. Solution structure and dynamics of the wild-type pseudoknot of human telomerase RNA. J Mol Biol. 2008;384:1249–61. doi: 10.1016/j.jmb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–15. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 54.Koirala D, Dhakal S, Ashbridge B, Sannohe Y, Rodriguez R, et al. A single-molecule platform for investigation of interactions between G-quadruplexes and small-molecule ligands. Nat Chem. 2011;3:782–87. doi: 10.1038/nchem.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lai CK, Miller MC, Collins K. Template boundary definition in Tetrahymena telomerase. Genes Dev. 2002;16:415–20. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lai CK, Miller MC, Collins K. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol Cell. 2003;11:1673–83. doi: 10.1016/s1097-2765(03)00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee JY, Kim DS. Dramatic effect of single-base mutation on the conformational dynamics of human telomeric G-quadruplex. Nucleic Acids Res. 2009;37:3625–34. doi: 10.1093/nar/gkp216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee JY, Okumus B, Kim DS, Ha T. Extreme conformational diversity in human telomeric DNA. PNAS. 2005;102:18938–43. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee JY, Yoon J, Kihm HW, Kim DS. Structural diversity and extreme stability of unimolecular Oxytricha nova telomeric G-quadruplex. Biochemistry. 2008;47:3389–96. doi: 10.1021/bi702013d. [DOI] [PubMed] [Google Scholar]
- 60.Lin J, Countryman P, Buncher N, Kaur PEL, et al. TRF1 and TRF2 use different mechanisms to find telomeric DNA but share a novel mechanism to search for protein partners at telomeres. Nucleic Acids Res. 2014;42:2493–504. doi: 10.1093/nar/gkt1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin J, Countryman P, Chen H, Pan H, Fan Y, et al. Functional interplay between SA1 and TRF1 in telomeric DNA binding and DNA–DNA pairing. Nucleic Acids Res. 2016;44:6363–76. doi: 10.1093/nar/gkw518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu S-W, Chu J-F, Tsai C-T, Fang H-C, Chang T-C, Li H-W. Assaying the binding strength of G-quadruplex ligands using single-molecule TPM experiments. Anal Biochem. 2013;436:101–8. doi: 10.1016/j.ab.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 63.Long X, Parks JW, Bagshaw CR, Stone MD. Mechanical unfolding of human telomere G-quadruplex DNA probed by integrated fluorescence and magnetic tweezers spectroscopy. Nucleic Acids Res. 2013;41:2746–55. doi: 10.1093/nar/gks1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Long X, Stone MD. Kinetic partitioning modulates human telomere DNA G-quadruplex structural polymorphism. PLOS ONE. 2013;8:e83420. doi: 10.1371/journal.pone.0083420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lue NF, Zhou R, Chico L, Mao N, Steinberg-Neifach O, Ha T. The telomere capping complex CST has an unusual stoichiometry, makes multipartite interaction with G-tails, and unfolds higher-order G-tail structures. PLOS Genet. 2013;9:e1003145. doi: 10.1371/journal.pgen.1003145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lynch S, Baker H, Byker SG, Zhou D, Sinniah K. Single molecule force spectroscopy on G-quadruplex DNA. Chemistry. 2009;15:8113–16. doi: 10.1002/chem.200901390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mason M, Schuller A, Skordalakes E. Telomerase structure function. Curr Opin Struct Biol. 2011;21:92–100. doi: 10.1016/j.sbi.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 68.McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–82. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mihalusova M, Wu JY, Zhuang X. Functional importance of telomerase pseudoknot revealed by single-molecule analysis. PNAS. 2011;108:20339–44. doi: 10.1073/pnas.1017686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miracco EJ, Jiang J, Cash DD, Feigon J. Progress in structural studies of telomerase. Curr Opin Struct Biol. 2014;24:115–24. doi: 10.1016/j.sbi.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, et al. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 72.Moffitt JR, Chemla YR, Smith SB, Bustamante C. Recent advances in optical tweezers. Annu Rev Biochem. 2008;77:205–28. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 73.Muller HJ. The remaking of chromosomes. Collect Net. 1938;13:182–98. [Google Scholar]
- 74.Müller S, Laxmi-Reddy K, Jena PV, Baptiste B, Dong Z, et al. Targeting DNA G-quadruplexes with helical small molecules. Chem Bio Chem. 2014;15:2563–70. doi: 10.1002/cbic.201402439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–59. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 76.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Noer SL, Preus S, Gudnason D, Aznauryan M, Mergny J-L, Birkedal V. Folding dynamics and conformational heterogeneity of human telomeric G-quadruplex structures in Na+ solutions by single molecule FRET microscopy. Nucleic Acids Res. 2016;44:464–71. doi: 10.1093/nar/gkv1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O’Donnell M, Li H. The eukaryotic replisome goes under the microscope. Curr Biol. 2016;26:R247–56. doi: 10.1016/j.cub.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oganesian L, Bryan TM. Physiological relevance of telomeric G-quadruplex formation: a potential drug target. Bio Essays. 2007;29:155–65. doi: 10.1002/bies.20523. [DOI] [PubMed] [Google Scholar]
- 80.Okamoto K, Sannohe Y, Mashimo T, Sugiyama H, Terazima M. G-quadruplex structures of human telomere DNA examined by single molecule FRET and BrG-substitution. Bioorg Med Chem. 2008;16:6873–79. doi: 10.1016/j.bmc.2008.05.053. [DOI] [PubMed] [Google Scholar]
- 81.Okumus B, Ha T. Real-time observation of G-quadruplex dynamics using single-molecule FRET microscopy. Methods Mol Biol. 2010;608:81–96. doi: 10.1007/978-1-59745-363-9_6. [DOI] [PubMed] [Google Scholar]
- 82.Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41:181–90. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 83.Parks JW, Kappel K, Das R, Stone MD. Single molecule FRET-Rosetta reveals RNA structural rearrangements during human telomerase catalysis. RNA. 2017;23:175–188. doi: 10.1261/rna.058743.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parks JW, Stone MD. Coordinated DNA dynamics during the human telomerase catalytic cycle. Nat Commun. 2014;5:4146. doi: 10.1038/ncomms5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qureshi MH, Ray S, Sewell AL, Basu S, Balci H. Replication protein A unfolds G-quadruplex structures with varying degrees of efficiency. J Phys Chem B. 2012;116:5588–94. doi: 10.1021/jp300546u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ray S, Bandaria JN, Qureshi MH, Yildiz A, Balci H. G-quadruplex formation in telomeres enhances POT1/TPP1 protection against RPA binding. PNAS. 2014;111:2990–95. doi: 10.1073/pnas.1321436111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ray S, Qureshi MH, Malcolm DW, Budhathoki JB, Celik U, Balci H. RPA-mediated unfolding of systematically varying G-quadruplex structures. Biophys J. 2013;104:2235–45. doi: 10.1016/j.bpj.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ren X, Li H, Clarke RW, Alves DA, Ying L, et al. Analysis of human telomerase activity and function by two color single molecule coincidence fluorescence spectroscopy. J Am Chem Soc. 2006;128:4992–5000. doi: 10.1021/ja056613z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rezler EM, Bearss DJ, Hurley LH. Telomeres and telomerases as drug targets. Curr Opin Pharmacol. 2002;2:415–23. doi: 10.1016/s1471-4892(02)00182-0. [DOI] [PubMed] [Google Scholar]
- 90.Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–37. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Robart AR, Collins K. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol Cell. 2011;42:308–18. doi: 10.1016/j.molcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–16. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–97. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sauerwald A, Sandin S, Cristofari G, Scheres SH, Lingner J, Rhodes D. Structure of active dimeric human telomerase. Nat Struct Mol Biol. 2013;20:454–60. doi: 10.1038/nsmb.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schmidt JC, Cech TR. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev. 2015;29:1095–105. doi: 10.1101/gad.263863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schmidt JC, Zaug AJ, Cech TR. Live cell imaging reveals the dynamics of telomerase recruitment to telomeres. Cell. 2016;166:1188–97. doi: 10.1016/j.cell.2016.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Selvam S, Koirala D, Yu Z, Mao H. Quantification of topological coupling between DNA super-helicity and G-quadruplex formation. J Am Chem Soc. 2014;136:13967–70. doi: 10.1021/ja5064394. [DOI] [PubMed] [Google Scholar]
- 98.Selvam S, Yu Z, Mao H. Exploded view of higher order G-quadruplex structures through click-chemistry assisted single-molecule mechanical unfolding. Nucleic Acids Res. 2016;44:45–55. doi: 10.1093/nar/gkv1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Selvin PR, Ha T. Single-Molecule Techniques: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 2008. [Google Scholar]
- 100.Sexton AN, Regalado SG, Lai CS, Cost GJ, O’Neil CM, et al. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev. 2014;28:1885–99. doi: 10.1101/gad.246819.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shim JW, Tan Q, Gu L-Q. Single-molecule detection of folding and unfolding of the G-quadruplex aptamer in a nanopore nanocavity. Nucleic Acids Res. 2009;37:972–82. doi: 10.1093/nar/gkn968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith FW, Feigon J. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature. 1992;356:164–68. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- 104.Songyang Z, Liu D. Inside the mammalian telomere interactome: regulation and regulatory activities of telomeres. Crit Rev Eukaryot Gene Expr. 2006;16:103–18. doi: 10.1615/critreveukargeneexpr.v16.i2.10. [DOI] [PubMed] [Google Scholar]
- 105.Stone MD, Mihalusova M, O’Connor CM, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–61. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17:671–82. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 107.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr Opin Struct Biol. 2006;16:307–18. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 108.Theimer CA, Finger LD, Trantirek L, Feigon J. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. PNAS. 2003;100:449–54. doi: 10.1073/pnas.242720799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tinoco I, Jr, Gonzalez RL., Jr Biological mechanisms, one molecule at a time. Genes Dev. 2011;25:1205–31. doi: 10.1101/gad.2050011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tippana R, Xiao W, Myong S. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014;42:8106–14. doi: 10.1093/nar/gku464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.van Oijen AM, Loparo JJ. Single-molecule studies of the replisome. Annu Rev Biophys. 2010;39:429–48. doi: 10.1146/annurev.biophys.093008.131327. [DOI] [PubMed] [Google Scholar]
- 112.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–31. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 113.Vulliamy TJ, Dokal I. Dyskeratosis congenita: the diverse clinical presentation of mutations in the telomerase complex. Biochimie. 2008;90:122–30. doi: 10.1016/j.biochi.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 114.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, et al. The POT1–TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–10. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 115.Wang H, Nora GJ, Ghodke H, Opresko PL. Single molecule studies of physiologically relevant telomeric tails reveal POT1 mechanism for promoting G-quadruplex unfolding. J Biol Chem. 2011;286:7479–89. doi: 10.1074/jbc.M110.205641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang Y, Patel DJ. Solution structure of a parallel-stranded G-quadruplex DNA. J Mol Biol. 1993;234:1171–83. doi: 10.1006/jmbi.1993.1668. [DOI] [PubMed] [Google Scholar]
- 117.Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 118.Wenz C, Enenkel B, Amacker M, Kelleher C, Damm K, Lingner J. Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 2001;20:3526–34. doi: 10.1093/emboj/20.13.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wolna AH, Fleming AM, Burrows CJ. Single-molecule analysis of thymine dimer-containing G-quadruplexes formed from the human telomere sequence. Biochemistry. 2014;53:7484–93. doi: 10.1021/bi501072m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wright WE, Shay JW. Cellular senescence as a tumor-protection mechanism: the essential role of counting. Curr Opin Genet Dev. 2001;11:98–103. doi: 10.1016/s0959-437x(00)00163-5. [DOI] [PubMed] [Google Scholar]
- 121.Wu JY, Stone MD, Zhuang X. A single-molecule assay for telomerase structure-function analysis. Nucleic Acids Res. 2010;38:e16. doi: 10.1093/nar/gkp1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu RA, Collins K. Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. EMBO J. 2014;33:921–35. doi: 10.1002/embj.201387205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wu RA, Dagdas YS, Yilmaz ST, Yildiz A, Collins K. Single-molecule imaging of telomerase reverse transcriptase in human telomerase holoenzyme and minimal RNP complexes. eLife. 2015;4:e08363. doi: 10.7554/eLife.08363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ying L, Green JJ, Li H, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. PNAS. 2003;100:14629–34. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.You H, Wu J, Shao F, Yan J. Stability and kinetics of c-MYC promoter G-quadruplexes studied by single-molecule manipulation. J Am Chem Soc. 2015;137:2424–27. doi: 10.1021/ja511680u. [DOI] [PubMed] [Google Scholar]
- 126.You H, Zeng X, Xu Y, Lim CJ, Efremov AK, et al. Dynamics and stability of polymorphic human telomeric G-quadruplex under tension. Nucleic Acids Res. 2014;42:8789–95. doi: 10.1093/nar/gku581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhou R, Zhang J, Bochman ML, Zakian VA, Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. eLife. 2014;3:e02190. doi: 10.7554/eLife.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]