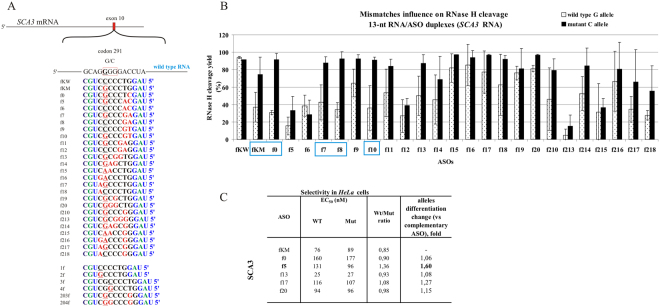
Figure 10.
Activity of gapmers designed to SCA3 RNA target in vitro and in HeLa cells. (A) Target SCA3 mRNA sequence containing site of G-to-C SNP and designed to it antisense gapmers. SNP in RNA strand is bolded and underlined, codon in which it occur is marked by red frame. Modified nucleotides within gapmers are marked by colours: blue-LNA, green-2′-O-MeRNA, bolded black- DNA, red-mismatched nucleotides, underlined-SNP site. (B) RNase H in vitro assay results for SCA3 RNA target, showing the influence of arrangements of mismatched nucleotides in 13 nt-long RNA/ASO duplexes on RNase H cleavage. Blue frames mark gapmers which cause preferential cleavage of Mut RNA. (C) Results of ASOs transfection to HeLa cells. The EC50 values for gapmers tested in presence of WT and mutant alleles were estimated based on dose-response curves fitted to experimental data in Origin Lab 8.0 software (for statistics see Supplementary data). The data were gathered from at least three separate experiments involving up to five concentrations in the range 10–100 nM. Comparison of EC50 values for gapmers between WT and Mutant target RNA alleles allowed to evaluate allele-preference for cleavage of tested gapmers. Two last columns of the table present WT/Mut ratio of gapmer selectivity and the selectivity of mismatched gapmers relative to complementary gapmer fKM.