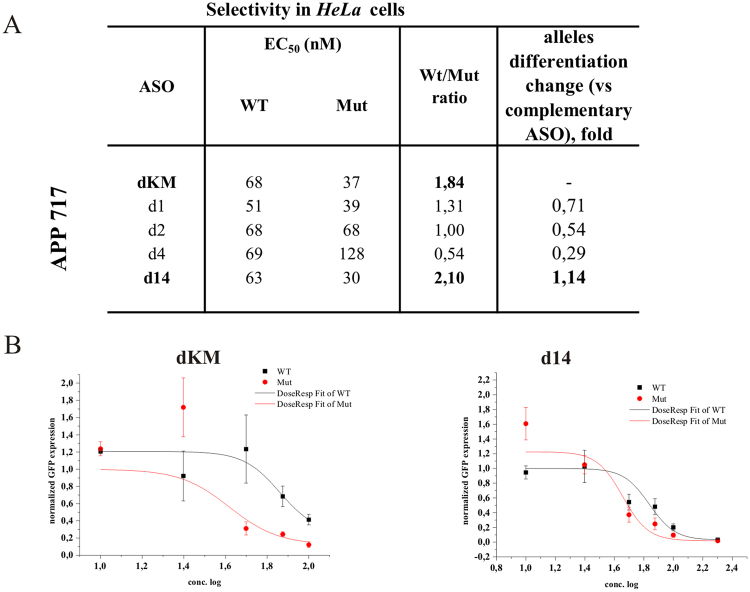
Figure 6.
Activity of selected gapmers, designed to APP 717 target, in HeLa cells. (A) Results of ASOs transfection to HeLa cells. The EC50 values for gapmers tested in presence of WT and mutant alleles were estimated based on dose-response curves fitted to experimental data in Origin Lab 8.0 software (for statistics see Supplementary data). The data were gathered from at least three separate experiments involving five concentrations in the range 10–100nM. Comparison of EC50 values for gapmers between WT and Mutant target RNA alleles allowed to evaluate allele-preference for RNA cleavage of tested gapmers. Two last columns of the table present WT/Mut ratio of gapmer selectivity and the selectivity of mismatched gapmers relative to complementary gapmer dKM. Plots present dose-response curves for selected antisense gapmers. (B) Dose-response curves for selected antisense gapmers. dKM - referenced ASO gapmer (mutant complementary to RNA717 target); d14 – ASO gapmer presenting SNP preference in HeLa cells, however not in the RNase H assay in vitro. Although the both models for d14 gapmer were statistically insignificant (P > 0.05), validation experiment confirmed SNP-preferential cleavage in concentrations determined from the curves (Fig. S13).