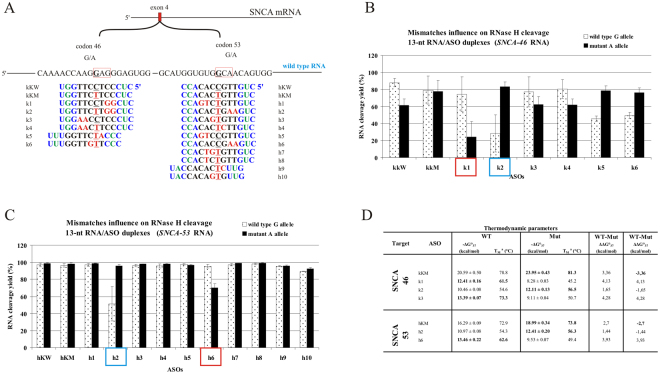
Figure 7.
Comprehensive results of in vitro activity of gapmers designed to SNCA RNA 46 and RNA 53 targets. (A) Target SNCA mRNA sequence containing sites of two G-to-A SNPs (RNA46 and RNA53) and designed to them antisense gapmers. SNPs in RNA strand are bolded and underlined, codons in which they occur are marked by red frame. Modified nucleotides within gapmers are marked by colours: blue-LNA, green-2′-O-MeRNA, bolded black- DNA, red-mismatched nucleotides, underlined-SNP site. (B) RNase H in vitro assay results for SNCA RNA46 target, showing the influence of arrangements of mismatched nucleotides in RNA/ASO duplexes on RNase H cleavage. Blue frames mark gapmers which cause selective cleavage of Mut RNA, red frames mark gapmers which preferentially cause wild type RNA cleavage. (C) RNase H in vitro assay results for SNCA RNA53 target, showing the influence of arrangements of mismatched nucleotides in RNA/ASO duplexes on RNase H cleavage. Blue frames mark gapmers which cause selective cleavage of Mut RNA, red frames mark gapmers which preferentially cause wild type RNA cleavage. (D) Thermodynamic parameters of SNCA wild type and mutants RNA/gapmer duplexes containing mismatches. Oligonucleotides, which differentiated two alleles cleavage yields in RNase H assay were only measured. Parameters for more stable duplex of the two (wild type/ASO or mutant/ASO) are bolded.