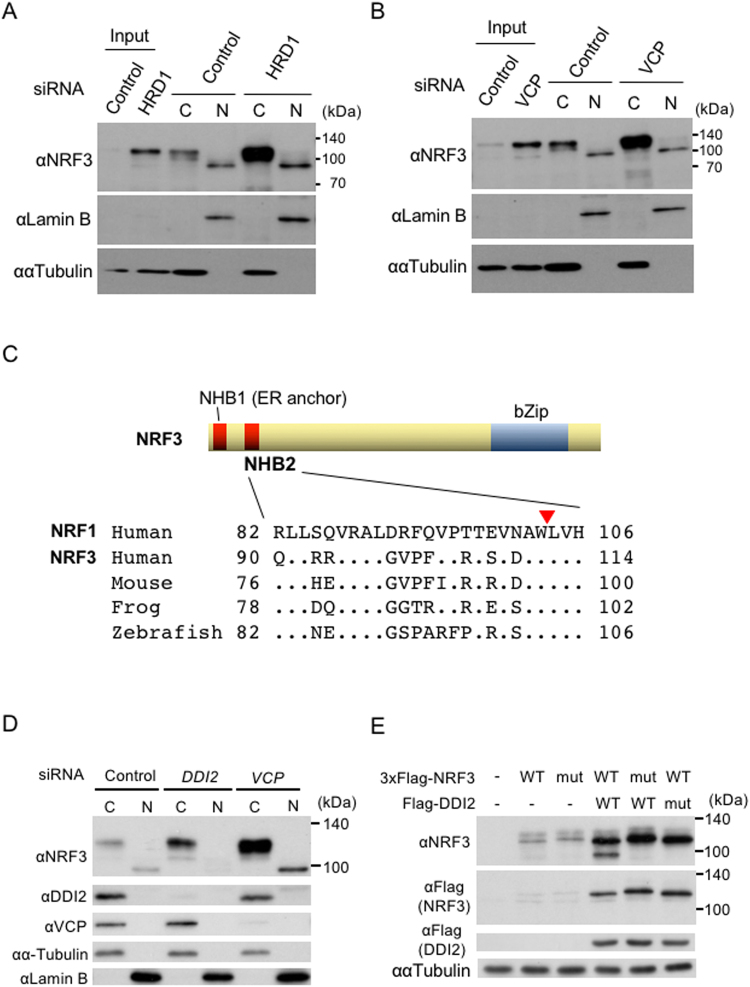
Figure 4.
The nuclear translocation of NRF3 requires the aspartic protease DDI2, but not inhibition of HRD1 or VCP. (A and B) HRD1 or VCP knockdown did not promote the nuclear translocation of NRF3. The DLD-1 cells were transfected with Control and HRD1 siRNA. At 48 hr after transfection, the cytoplasmic and nuclear fractions were extracted from the cells and subjected to immunoblot analysis with anti-NRF3 antibody. Lamin B and α-Tubulin were utilized as the nuclear and cytoplasmic markers, respectively. (C) Sequence alignment of the NHB2 domain of NRF1 and NRF3. The NRF1 processing site (a red triangle)27 is highly conserved in NRF3 among several species. (D) DDI2 knockdown substantially abolishes the nuclear translocation of the endogenous NRF3 in DLD-1 cells. After transfection with the indicated siRNA, the cells were fractionated into the cytoplasmic (C) and nuclear extracts (N), followed by an immunoblot analysis using the indicated antibodies. As a positive control, a similar experiment using VCP siRNA was performed. (E) DDI2 cleaves the N-terminal 3xFlag-fused hNRF3. The cells were transfected into HeLa cells with the indicated plasmids, and whole-cell extracts from the cells were subjected to immunoblot analysis with the indicated antibodies. 3xFlag-hNRF3 WL111AA (mut) is the mutant of a putative cleavage site in NRF3 that corresponds to the same site in NRF127, and DDI2 D252N (mut) is the protease dead mutant28.