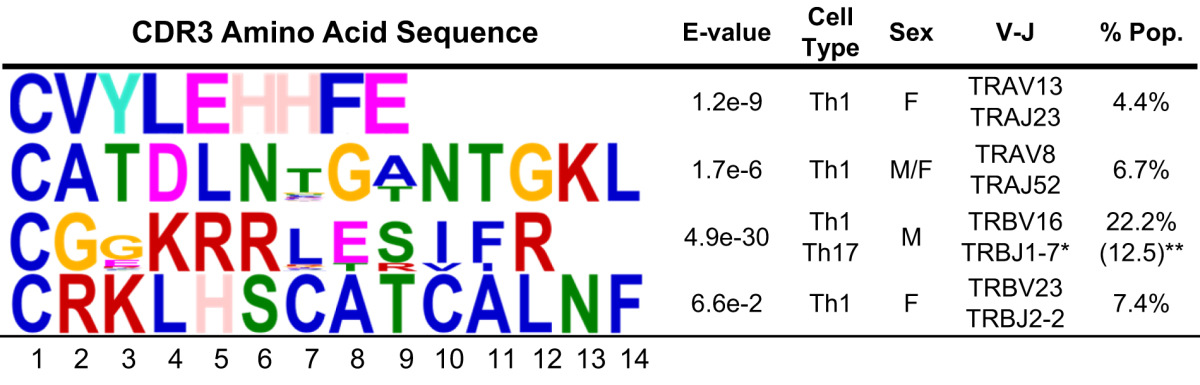
Table 2.
MEME analysis of high frequency CDR3 regions from SjSS mice. Amino acid motif analysis was performed on the high frequency CDR3 sequences from the B6.NOD-Aec1/2 mice. Bit height corresponds to the likelihood of the amino acid in each position. Blue – hydrophobic, neutral amino acids, Red – positively charge hydrophilic amino acids, Green – Neutral hydrophilic amino acids, Magenta – negatively charged hydrophilic amino acids, Orange-glycine, Teal-Tyrosine, Pink-Histidine (positively charged moderately hydrophobic). E-value indicates the model confidence for the amino acid in that position. Cytokine indicates the secreted molecule(s) associated with the motif, V/J indicates recombination gene segment pairings, and % pop. Indicates the percent of the motif present in the IFN-γ or the IL-17A producing population. *Most popular V/J combination. **Motif located in both IL-17A and IFN-γ producing populations in both TCRα and TCRβ chains.