Abstract
The centrosome serves as a major microtubule-organizing center (MTOC). The Cdc6 protein is a component of the pre-replicative complex and a licensing factor for the initiation of chromosome replication and localizes to centrosomes during the S and G2 phases of the cfell cycle of human cells. This cell cycle-dependent localization of Cdc6 to the centrosome motivated us to investigate whether Cdc6 negatively regulates MTOC activity and to determine the integral proteins that comprise the pericentriolar material (PCM). Time-lapse live-cell imaging of microtubule regrowth revealed that Cdc6 depletion increased microtubule nucleation at the centrosomes and that expression of Cdc6 in Cdc6-depleted cells reversed this effect. This increase and decrease in microtubule nucleation correlated with the centrosomal intensities of PCM proteins such as γ-tubulin, pericentrin, CDK5 regulatory subunit-associated protein 2 (CDK5RAP2), and centrosomal protein 192 (Cep192). The regulation of microtubule nucleation and the recruitment of PCM proteins to the centrosome required Cdc6 ATPase activity, as well as a centrosomal localization of Cdc6. These results suggest a novel function for Cdc6 in coordinating centrosome assembly and function.
Keywords: ATPases associated with diverse cellular activities (AAA), cell cycle, centrosome, DNA replication, microtubule
Introduction
The centrosome, which plays roles in diverse cellular and developmental processes, consists of a pair of centrioles surrounded by pericentriolar material (PCM)3 (1–3). PCM is not a membrane-bound, amorphous mass, but rather contains a highly ordered hierarchical organization. The integral scaffold proteins, such as pericentrin, CDK5RAP2, Cep192, Cep152, and Nedd1, exhibit concentric toroidal distributions in interphase PCM (4, 5). PCM is matured and expanded at the onset of mitosis to form bipolar spindles for chromosome segregation in mitosis. During interphase, centrosomes associated closely with the nucleus are duplicated; they then separate and migrate to the poles of the dividing cell, to function as spindle poles during mitosis (6).
The centrosome serves as the major microtubule-organizing center (MTOC). Microtubules are nucleated at the γ-tubulin ring complexes (γ-TuRC) embedded in the PCM (7–9). γ-Tubulin in the γ-TuRC functions as a nucleation core for microtubule polymerization. α- and β-tubulins polymerize into microtubules (10–12). Tubulin modifications and microtubule-associated proteins (MAPs) regulate microtubule dynamics (13, 14). During interphase, microtubules originating from centrosomes participate in positioning the nucleus (15). Microtubules play important roles in diverse cellular functions, including chromosome segregation, vesicular transport, cell motility, cell shape, and motility (16–19).
To initiate eukaryotic chromosome replication during the G1 phase of the cell cycle, the association of Cdc6 with an origin recognition complex (ORC), bound to the origin of replication, recruits Cdt1–MCM2–7 complexes to form the pre-replicative complex (pre-RC) (20, 21). Cyclin A/Cdk2 phosphorylation and nuclear export signals promote non-chromatin bound Cdc6 to translocate to the cytoplasm during the G1/S transition phase (22–24). Pre-RC formation during the G1 phase is essential to ensuring that chromosomal replication occurs only once per cell cycle. As a member of the AAA+ family of ATPases, Cdc6 possesses the Walker A phosphate-binding loop (P-loop) and the Walker B motif, both of which are essential for ATP binding and hydrolysis. Cdc6 is highly conserved within metazoans; its ATPase activity is required for pre-RC assembly and for other biological functions (25–28). p21 and p27 bound to Cdk2 are eliminated by Cdc6 to activate Cdk2 for cell cycle progression (29, 30). Studies have also shown that the formation of stable complexes of Cdc6 with Apaf-1, activated by cytochrome c, inhibit the apoptosome assembly required for cell death (31).
Although pre-RC formation occurs during G1 phase, Cdc6 protein levels are minimal during this phase, and its mRNA and protein levels do not begin to increase until S phase (32–34). Non-chromatin bound Cdc6 is exported from the nucleus to the cytoplasm during S phase (32, 35). Furthermore, Cdc6 localizes to centrosomes during both the S and G2 phases (36, 37). In this work, we address Cdc6 function at centrosomes during the S and G2 phases.
Results
Cdc6 depletion increases microtubule regrowth originating from centrosomes
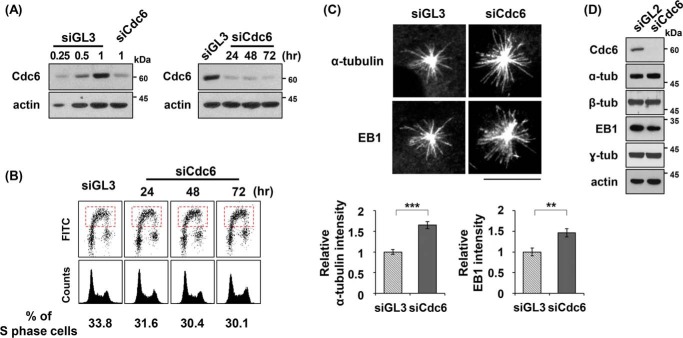
Cdc6 localizes to the centrosomes during the S and G2 phases of the human cell cycle (supplemental Fig. S1, A and B) (36, 37). Because the centrosome functions as a MTOC, we examined whether Cdc6 plays a role in microtubule formation. siRNA-mediated partial depletion of Cdc6 in U2OS cells was performed under conditions that minimized its incidental influence on chromosome replication and cell cycle progression (Fig. 1, A and B). Partial depletion with a Cdc6-specific siRNA for 24 h caused cells to contain ∼25% of the Cdc6 protein amounts seen in cells treated with control siRNA GL3. This partial depletion also marginally affected DNA synthesis, as determined by BrdU incorporation followed by fluorescence-activated cell sorting (FACS) analysis.
Figure 1.
Cdc6 depletion increases microtubule regrowth from centrosomes. A, asynchronously grown U2OS cells were transfected with control GL3 or Cdc6-specific siRNA (see “Experimental procedures”) for 24 h or the indicated times, and then subjected to immunoblot analysis. The lysates of the control siRNA-treated cells were loaded with the indicated, relative volumes. Actin served as an internal control. B, FACS analysis was performed with BrdU (lower top) and propidium iodide (lower bottom) staining of the Cdc6-depleted cells for the indicated times after transfection. S phase cells are indicated in the dashed boxes. The proportions of replicating S phase cells are shown below the FACS profiles. siGL3, control siRNA GL3; siCdc6, Cdc6-specific siRNA. C, microtubule regrowth assays were performed with cells treated with the indicated siRNA for 24 h, as described under “Experimental procedures.” The cells, after incubation on ice for 1 h to depolymerize the microtubules, were incubated in a fresh medium at 37 °C for 15 s, and then fixed in the PEM + Fixative buffer. Microtubules were immunostained with antibodies specific to α-tubulin or EB1. Centrosomal intensities of α-tubulin and EB1 were densitometrically determined, and relative fluorescent intensities of α-tubulin and EB1 were plotted. Values represent mean ± S.D. of at least 100 cells in each of three independent experiments (**, p < 0.01; ***, p < 0.001). Scale bar, 10 μm. D, the indicated proteins were detected in immunoblots with antibodies specific to each protein.
Cdc6-depleted cells were incubated on ice for 1 h to depolymerize microtubules, followed by incubation at 37 °C to allow microtubule regrowth (38). Microtubule regrowth from the centrosomes was measured for 15 s by immunostaining cells with either anti-α-tubulin or anti-EB1 antibodies to detect microtubule formation (Fig. 1C). α-Tubulin is a structural subunit of microtubules (12). EB1, which is a microtubule plus-end tracking protein (39), produces “comets” that indicate microtubules emanating from the centrosome (40). Although the microtubule regrowth occurred in a time-dependent manner, the rate of microtubule regrowth at 37 °C appeared to decrease after 10 s of incubation (supplemental Fig. 2A). The intensities of both α-tubulin and EB1 were significantly higher in Cdc6-depleted cells than in control cells (Fig. 1C and supplemental Fig. 2A). Depletion of Cdc6 did not significantly influence the amounts of α-tubulin, β-tubulin, or other proteins that are involved in microtubule formation (Fig. 1D). Therefore, the increased microtubule regrowth in Cdc6-depleted cells suggests that Cdc6 inhibits microtubule formation, rather than affecting (perhaps by down-regulating) the expression of the proteins that participate in microtubule formation.
Centrosomal localization of Cdc6 protein reduces microtubule formation
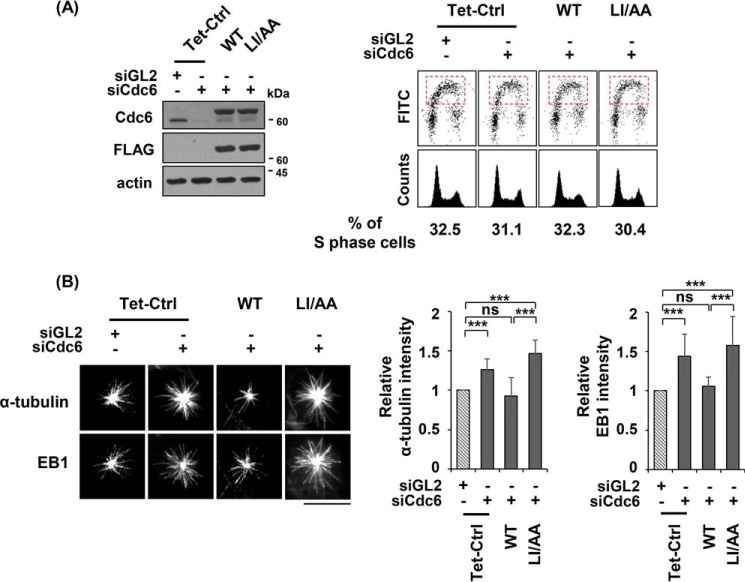
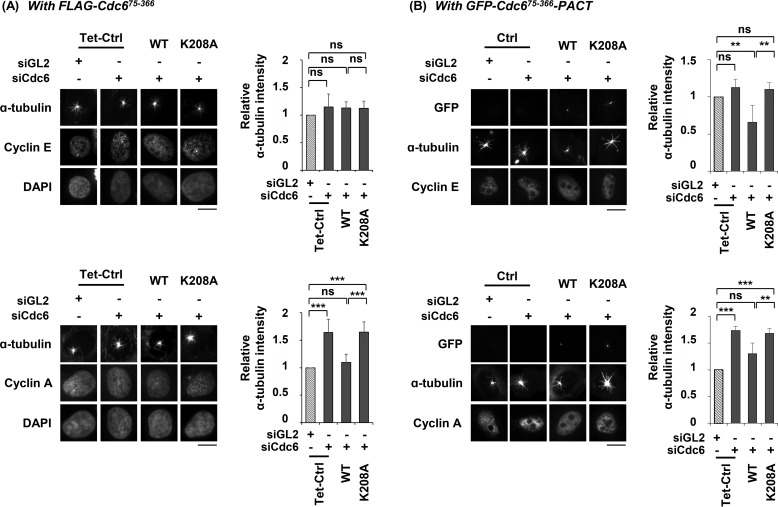
We determined whether centrosomal localization of Cdc6 is required to inhibit microtubule formation. Amino acid residues 311–366 of Cdc6 were determined to be the centrosomal localization signal (CLS) region of the protein (therefore indicated as Cdc6-CLS) (36). The Cdc6-CLS allowed fused tag proteins, such as DsRed, GFP, and FLAG, to locate at the centrosome. Substitutions of Leu-313, Ile-316, or both, which are highly conserved within metazoans, with Ala prevented localization of the substituted Cdc6 to the centrosome (36) (supplemental Fig. S1C). Cdc6(LI/AA) contained substitutions of both Leu-313 and Ile-316 with Ala. Cdc6 siRNA-resistant, [FLAG-Cdc6 wild type] or [FLAG-Cdc6(LI/AA)] coding sequences were introduced into a U2OS Tet-On cell line. Addition of doxycycline to the cultures induced expression of the corresponding Cdc6 protein (Fig. 2A). Induction of wild-type or mutant protein expression in Cdc6-depleted cells did not significantly alter the cell cycle progression. Cdc6 depletion increased microtubule regrowth detected by α-tubulin and EB1 intensities, as shown in Figs. 1B and 2B. Whereas induction of wild-type Cdc6 in Cdc6-depleted cells reduced α-tubulin and EB1 intensities, the CLS-mutant Cdc6(LI/AA), which was defective in centrosomal localization, did not display these reductions (Fig. 2C and supplemental Fig. S2B). The inability of Cdc6(LI/AA) to rescue the Cdc6 depletion suggests that Cdc6 needs to locate at centrosomes to reduce microtubule formation.
Figure 2.
Centrosomal localization of Cdc6 is required to reduce microtubule formation. A, asynchronously grown U2OS Tet-On cells expressing Cdc6-siRNA resistant FLAG-Cdc6 wild type or FLAG-Cdc6(LI/AA) (see “Experimental procedures”) were transfected with the indicated siRNA for 24 h. The proteins were induced by addition of 2 μg/ml of doxycycline, 24 h prior to siRNA treatment. The indicated proteins were detected by immunoblotting. Replicating S phase cells are indicated in dashed boxes. Proportions of replicating S phase cells are shown below the FACS profiles. B, microtubule regrowth assays with incubation at 37 °C for 15 s, after incubation on ice for 1 h to depolymerize the microtubules, and then fixed in the PEM + Fixative buffer. Quantification and statistical analyses were performed as described in the legend to Fig. 1C. Values represent mean ± S.D. of at least 30 cells in each of the three independent experiments (ns, not significant; ***, p < 0.001). Scale bar, 10 μm; Tet-Ctrl, control U2OS Tet-On cells; WT, FLAG-Cdc6 wild type; LI/AA, FLAG-Cdc6(L313A/I316A); siGL3, control siRNA GL3; siCdc6, Cdc6-specific siRNA.
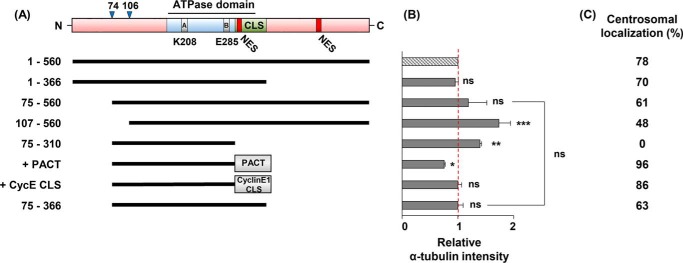
To determine the region(s) of the Cdc6 protein participating in the inhibition of microtubule formation, U2OS cells were transfected with deletion constructs of Cdc6 (Fig. 3A). Endogenous Cdc6 protein within transfected cells was depleted using Cdc6-specific siRNA; this was followed by a microtubule regrowth assay for detecting α-tubulin (Fig. 3B). Centrosome localization of each Cdc6 deletion construct was detected via fluorescent microscopy (Fig. 3C, supplemental Figs. S1, B and D, and S3A). Although deletion of the C-terminal 367–560 or the N-terminal 1–74 amino acid residues reduced α-tubulin intensity, further deletions of the N terminus or C terminus residues had a negative effect on this α-tubulin intensity reduction. Fragment 75–366, which was generated by deletion of both the C-terminal 367–560 and the N-terminal 1–74 amino acid residues, maintained all the inhibitory activity of the full-length protein. Because fragment 75–310, which did not contain Cdc6-CLS, could not localize to the centrosome, this deletion mutant lacked the inhibitory effect. However, attaching either the PACT domain (amino acid residues 3699–3790) or the CycE-CLS (amino acid residues 231–250), which promotes the centrosomal localization of AKAP-450 (41) and Cyclin E (42), respectively, onto fragment 75–310 restored the inhibitory activity. These results suggest that fragment 75–310, upon localization to the centrosome, could reduce microtubule formation; furthermore, they indicated that this reduction in microtubule formation required the localization of Cdc6 to the centrosome.
Figure 3.
Amino acid residues 75–366 of Cdc6 participate in the reduction of microtubule formation. A, schematic structures of Cdc6 motifs and domains as shown previously (66) are described (top). Numbers represent the amino acid residues. The Ser residues 74 and 106, phosphorylation sites by CDKs; A, Walker A motif; B, Walker B motif; NES, nuclear export signal; PACT, PACT domain of AKAP-450; CycE-CLS, cyclin E1-CLS of Cyclin E. B, at 24 h after transfection with each DNA construct expressing the indicated fragments fused to the C termini of FLAG tag, U2OS cells were treated with Cdc6-specific siRNA; microtubule regrowth assays with incubation at 37 °C for 15 s were performed 24 h after siRNA treatment. Centrosmal α-tubulin intensities were quantified. C, centrosomal localization of each construct is shown in supplemental Fig. S1. Immunoblot of the exogenously expressed Cdc6 proteins is shown in supplemental Fig. S3A.
ATPase activity of Cdc6 is necessary for the reduction of microtubule formation
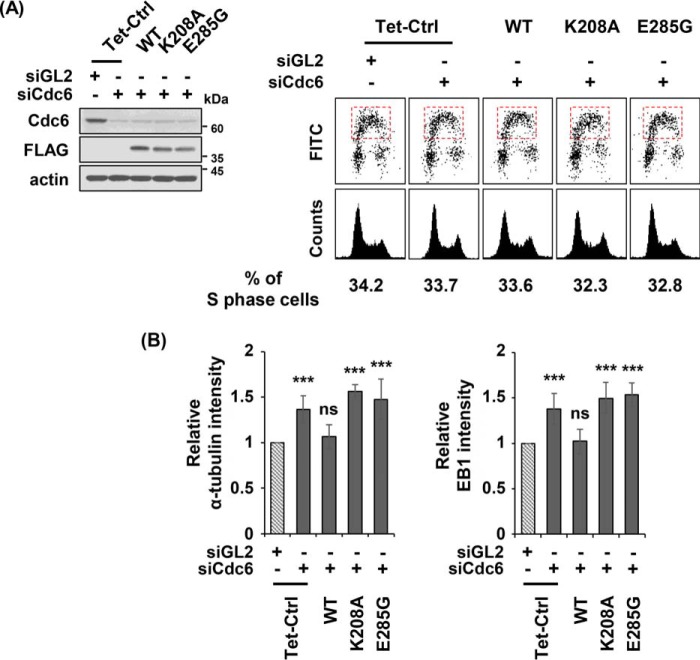
Cdc6(75–366), the 75–366 fragment of Cdc6, possesses an ATPase domain that harbors the Walker A motif or P-loop (amino acid residues 202–209), and the Walker B motif (284–287); these are essential for ATP binding and hydrolysis, respectively (Fig. 3A) (26). Both activities are indispensable for the role of the Cdc6 protein in the initiation of chromosome replication (26–28). Substitutions of an amino acid residue on either the Walker A or Walker B motif, which were accomplished via substitution of either Lys-208 or Glu-285 with Ala or Gly, respectively, were introduced into FLAG-tagged Cdc6(75–366). U2OS inducible Tet-On cell lines were then generated in which either the Cdc6 siRNA-resistant FLAG-Cdc6(75–366) wild-type (WT), Walker A (K208A), or Walker B (E285G) mutant proteins could be induced. The in vitro ATP hydrolytic activities of the Walker A and B mutant proteins were near background levels, but the activity of the CLS mutant protein Cdc6(LI/AA) was comparable to the corresponding wild-type protein (supplemental Figs. S3B and S4). Under conditions of endogenous Cdc6 depletion, induction of Cdc6(75–366) containing either the K208A or E285G substitutions did not significantly influence cell cycle progression when compared with the induction of the wild-type Cdc6(75–366) (Fig. 4A). Although the induced wild-type Cdc6(75–366) restored the inhibition of microtubule regrowth, the corresponding Walker A and Walker B mutants failed to accomplish this (Fig. 4B). The inactivity of the Walker A and Walker B mutant Cdc6(75–366) suggested that both the ATP-binding and hydrolysis activities of Cdc6 are involved in the reduction of microtubule formation.
Figure 4.
Both ATP-binding and hydrolysis activities of Cdc6 are required to reduce microtubule formation. A, depletions of endogenous Cdc6 and inductions of the indicated FLAG-Cdc6(75–366) in asynchronously grown U2OS Tet-On cell lines were performed as described in the legend to Fig. 2A. Immunoblot (left) and FACS (right) analyses were also performed as described in the legend to Fig. 2A. B, microtubule regrowth assay, with incubation at 37 °C for 15 s, was performed for at least 100 cells in each of the three independent experiments, as described in the legend to Fig. 1B. Tet-Ctrl, U2OS Tet-On control cells; WT, FLAG-Cdc6(75–366) wild-type: K208A, FLAG-Cdc6(75–366)(K208A); E285G, FLAG-Cdc6(75–366) (E285G).
Cdc6 reduces microtubule formation in S and G2 phase centrosomes
We determined whether the reduction of microtubule formation by Cdc6 is in concert with the cell-cycle dependent localization of Cdc6 to the centrosome during the S and G2 phases. Cyclin E is a marker of the G1 phase, and cyclin A is of both the S and G2 phases (43, 44). In Cdc6-depleted G1 phase cells that were detected by cyclin E-positive immunostaining, the differences between the induced FLAG-Cdc6(75–366) wild type and the corresponding Walker A mutant K208A, as well as between the control and Cdc6-depleted cells, were statistically insignificant in the microtubule regrowth assays (Fig. 5A, top). This lack of difference could be explained by the absence of Cdc6 at the G1-phase centrosome (supplemental Fig. S1B). In contrast, the induced wild-type protein decreased microtubule regrowth in S and G2 phase cells (Fig. 5A, bottom). Because Walker B mutant E285G behaved similarly to Walker B mutant K208A in the microtubule regrowth assay and in other assays described here, we did not include the results obtained with the K208A mutant in the following results.
Figure 5.
Cdc6 negatively regulates microtubule formation in S and G2 phase centrosomes. Microtubule regrowth assays with incubation at 37 °C for 15 s were performed on asynchronously grown cells. Cells were then immunostained with anti-cyclin E antibodies (top panel) or anti-cyclin A antibodies (bottom panel). Induction of the indicated Cdc6 proteins and detection and quantification of relative α-tubulin intensities were carried out as described in the legend to Fig. 4. A, U2OS Tet-On inducible cell lines used are described in the legend to Fig. 4. Tet-Ctrl, U2OS Tet-On control cells; WT, FLAG-Cdc6(75–366) wild type: K208A, FLAG-Cdc6(75–366)(K208A). B, FLAG-Cdc6(75–366)-PACT wild-type (WT) or Walker A mutant protein (K208A) was induced in FRT/TO HeLa cell lines.
Fusion of the PACT domain onto a protein causes the fused protein to localize to the centrosomes throughout the cell cycle (41). The fusion of the PACT domain to Cdc6(75–366) allowed it to localize to the G1 phase centrosomes, in contrast to the Cdc6(75–366) not fused to the PACT domain (supplemental Fig. S1B). The induced FLAG-Cdc6(75–366)-PACT wild-type protein reduced microtubule regrowth, but the corresponding Walker A mutant did not (Fig. 5B, top panel). These results imply that the Cdc6-dependent inhibition of microtubule formation, which requires centrosomal localization, occurs during the S and G2 phases.
Cdc6 controls microtubule nucleation
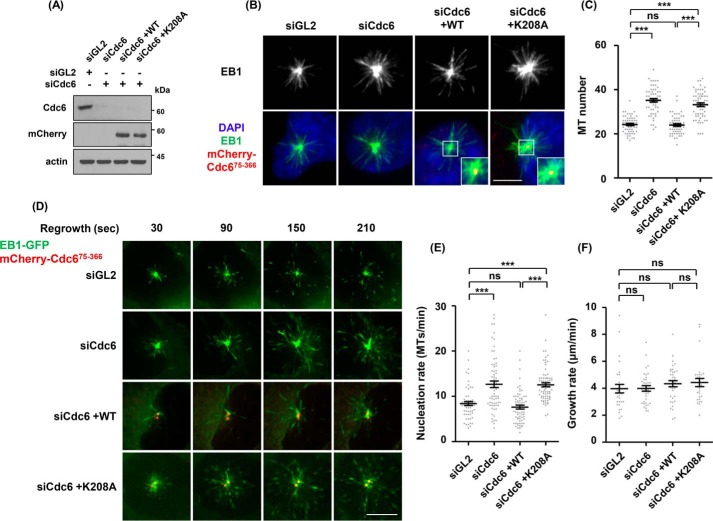
The association of the EB1 protein with growing microtubule tips (39) suggested that the increase and decrease of the intensity of EB1 shown in Figs. 1, 2, and 4 might have been caused by changes in the number of microtubules through Cdc6-mediated control of microtubule nucleation. Therefore, the number of microtubules originating from the centrosome was measured by counting EB1 comet numbers (Fig. 6, A–C). Depletion of Cdc6 resulted in increased microtubule numbers. Transfection of an mCherry-tagged Cdc6(75–366) wild-type-expressing DNA construct into Cdc6-depleted cells decreased the microtubule numbers compared with those of control GL2 siRNA-treated cells, whereas the corresponding Walker A mutant did not show this effect.
Figure 6.
Cdc6 controls microtubule nucleation. A–C, U2OS cells were treated with control siRNA (siGL2) or Cdc6-specific siRNA (siCdc6). Cdc6-depleted cells were transfected with mCherry-Cdc6(75–366) wild-type (siCdc6+WT)- or Walker A mutant (siCdc6+K208A)-expressing DNA constructs. A, immunoblot analyses were performed with the indicated antibodies. B, microtubule regrowth assay was performed at 37 °C for 15 s. The number of EB1 comets (green) immunostained with anti-EB1 antibody and fluorescence of mCherry-Cdc6 (red) were detected using DeltaVision. C, fields containing centrosomes are shown at higher magnifications in insets. The number of EB1 comets emanated from centrosomes of 30 cells per each treatment was quantified. D–F, the indicated DNA constructs were transfected into EB1-GFP-expressing U2OS stable cells as described in A. D, to perform live-cell time-lapse imaging of microtubule regrowth, depolymerized cells on ice were transferred to the chamber (Delta Vision) and prewarmed at 25 °C. Microtubule regrowth was performed at 25 °C, as described under “Experimental procedures.” Single frames of video for 10 cells per each treatment were collected at 30-s intervals (supplemental Movies A–D). Representative images are displayed. E, rates of microtubule nucleation were determined at 30-s intervals for 4 min by counting EB1-GFP comets emerging from the centrosome. F, rates of microtubules growth were quantified by measuring the length of each EB1-GFP comet from centrosomes. The length was manually tracked using SoftWoRx. At least 5 microtubules at early times of regrowth were tracked in 10 cells. Scatter plots were drawn using GraphPad software. ns, not significant; ***, p < 0.001; scale bar, 10 μm.
We carried out time-lapse imaging of live cells on the GFP-tagged EB1-expressing U2OS cell line. To slow down microtubule formation for live-cell imaging, microtubule regrowth assays were performed at 25 °C instead of 37 °C, which was used in the previous assays, after incubation on ice. The number of microtubules was determined by counting GFP fluorescent comets of EB1-GFP (Fig. 6, D and E, and supplemental Movies A–D). Cdc6 depletion increased microtubule numbers as shown in Fig. 6, B and C. Transfection of mCherry-Cdc6(75–366) wild-type expressing the DNA construct into the Cdc6-depleted cells significantly reduced microtubule numbers, but the corresponding Walker A mutant did not. In contrast, neither Cdc6 depletion nor transfection of the Cdc6(75–366) wild-type-expressing construct significantly changed the rates of microtubule growth (Fig. 6F). The increase of microtubule numbers by Cdc6 depletion, together with the reduction by Cdc6(75–366) expression in Cdc6-depleted cells, suggests that Cdc6 negatively controls microtubule nucleation at the centrosome.
Cdc6 negatively regulates the levels of PCM proteins at the centrosome
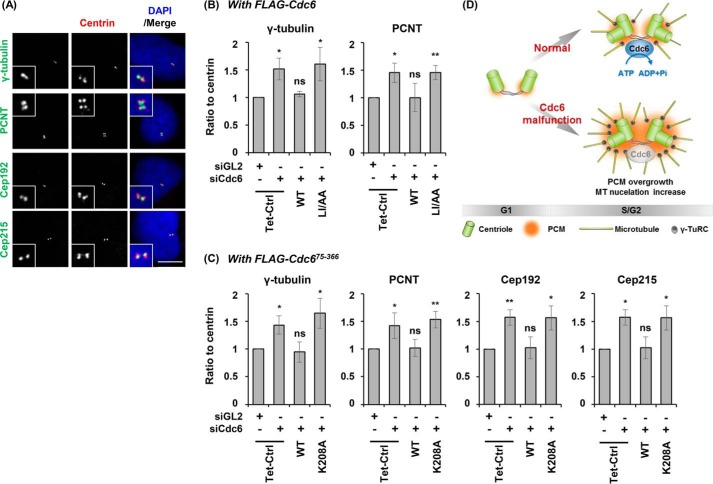
Because the centrosomal localization of Cdc6 negatively regulates microtubule nucleation, we investigated whether Cdc6 could also affect γ-tubulin levels at the centrosome. γ-Tubulin of the γ-TuRC functions as a nucleation core for microtubule polymerization (7–9). Because the depletion or induction of Cdc6 did not significantly affect the centrosomal intensities of centrin, which is located in the centrioles and the pericentriolar lattice of the centrosome (supplemental Fig. S5, A and B), the intensities of PCM proteins at centrosomes were compared with those of centrin (Fig. 7). Cdc6 depletion increased the intensity of γ-tubulin at the centrosomes (Fig. 7, A and B). This increase was reduced by the induction of the full-length wild-type Cdc6 protein, but not by the induction of Cdc6(LI/AA), which is defective in centrosomal localization (supplemental Fig. S1C). Similarly, Cdc6 depletion increased, whereas induction of the wild-type protein decreased, the intensity of pericentrin, a scaffold protein that exists within the PCM (45). Cdc6-depleted cells showed pronounced increases in γ-tubulin and pericentrin at centrosomes during the S and G2 phases (supplemental Fig. S5, C–E), supporting that Cdc6 functions at the centrosomes during the S and G2 phases.
Figure 7.
Cdc6 regulates the amounts of PCM proteins at the centrosome. Asynchronously grown U2OS Tet-On inducible cell lines used and induction of the indicated proteins are shown in Figs. 2 and 4. A, cells were co-immunostained with the indicated antibodies. Fields around the centrosomes containing 4 dots of centrin are shown at higher magnifications in insets. Scale bar, 10 μm. B, ratios are described as relative intensities of the indicated protein to relative intensity of centrin. PCNT, pericentrin; Tet-Ctrl, control U2OS Tet-On cells; WT, FLAG-Cdc6 full-length wild type; LI/AA, FLAG-Cdc6(LI/AA). C, Tet-Ctrl, U2OS Tet-On control cells; WT, FLAG-Cdc6(75–366) wild type; K208A, FLAG-Cdc6(75–366)(K208A). Values represent mean ± S.D. of at least 50 cells in three independent experiments. D, Cdc6 negatively controls PCM assembly and microtubule nucleation.
The control of pericentrin levels by Cdc6 led us to examine whether Cdc6 controls the levels of two other PCM scaffold proteins, Cep215 (also known as CDK5RAP2) and Cep192, at the centrosome. As seen with full-length Cdc6 (Fig. 7B), induction of Cdc6(75–366) wild-type protein reduced the intensities of γ-tubulin and pericentrin (Fig. 7C). Whereas depletion of Cdc6 increased the intensities of those PCM proteins, induction of Cdc6(75–366) wild-type reduced their intensities. However, the corresponding Walker A mutant, K208A, failed to reduce these PCM protein intensities. Pericentrin, CDK5RAP2, and Cep192 have been shown to be involved in the recruitment of γ-TuRC to the centrosome (4, 46–49). The increased levels of these PCM scaffold proteins and γ-tubulin caused by Cdc6 depletion, and conversely the reduction of their levels by the presence of Cdc6, together suggest that Cdc6 negatively controls recruitment of PCM proteins to the centrosomes and thereby modulates nucleation for microtubule formation (Fig. 7D).
Discussion
Although Cdc6 participates in pre-RC formation in the nucleus during the G1 phase, its protein levels are lowest during the G1 phase, and its mRNA and protein levels begin to increase from S phase (32–34). In addition, non-chromatin-bound Cdc6 is exported from the nucleus during the G1/S transition (22). These results suggest non-nuclear roles for Cdc6 in the S and G2 phases. Cdc6 localizes to centrosomes during the S and G2 phases (36, 37). This work demonstrates a novel function for Cdc6 at S and G2 phase centrosomes: that Cdc6, with its ATPase activity, negatively controls both microtubule nucleation for MTOC activity and the levels of PCM proteins (Fig. 7D). MTOC regulation appears to be achieved via the modulation of PCM protein levels at the centrosomes.
Many centrosomal proteins are involved in promoting MTOC activity, but only a few have been identified as negative regulators of the MTOC prior to mitosis. BRCA1 protein localized to the centrosome decreases microtubule nucleation by ubiquitinating Lys-344 of γ-tubulin (50). NEK2 phosphorylation of centrobin reduces microtubule networks during the G2 phase, whereas PLK1 phosphorylation increases microtubule stability for bipolar spindle formation during the M phase (51). In this study, we add that Cdc6 is a negative regulator of MTOC in interphase centrosomes. The functions of Cdc6 may differ from those of BRCA1 and centrobin, in that Cdc6 regulates the recruitment of the PCM proteins participating in MTOC assembly or activity.
The PCM in interphase centrosomes is organized in an orderly manner, with a concentric toroidal structure (4, 5). Pericentrin is an elongated molecule extending away from the centriole, whereas other PCM proteins, such as CDK5RAP2, Cep192, and NEDD1, occupy separable concentric domains surrounding the centrioles. As the ATPase activity of Cdc6 in protein complexes participates in modulating the functions of proteins within the complex (25–31), Cdc6 at the centrosome may form complexes with regulatory proteins to control PCM assembly. The increased intensities of centrosomal pericentrin, CDK5RAP2, and Cep192 caused by Cdc6 depletion (Fig. 7), along with the centrosomal localization and function of Cdc6 (Figs. 5 and 6), suggest that Cdc6 is an upstream regulator of PCM organization for maintenance of PCM proteins during interphase. As a result, the γ-TuRC may be limited to be recruited to the centrosome, in the presence of Cdc6, during S and G2 phase cells. Delocalization of Cdc6 from the centrosomes during mitosis temporally coincides with the maturation and expansion of the PCM at the onset of mitosis (36, 46).
Centrosome duplication and chromosomal replication share similarities in the following aspects. Both occur once per cell cycle, and in a cell cycle-dependent manner; these cell cycle-dependent processes are commonly regulated by cyclin-dependent kinases; and the duplicated centrosomes and chromosomes are equally segregated into daughter cells during mitosis (52–55). Furthermore, the pre-RC forming and controlling proteins, such as origin recognition complex subunits (56–58), MCM subunits (59, 60), and geminin (61), also exist in centrosomes to maintain centrosomal integrity. Our results show that Cdc6, which is a component of the pre-RC, localizes to centrosomes in a cell cycle-dependent manner, and that it modulates both the levels of PCM proteins and MTOC activity. The replicative proteins that regulate both the pre-RC and centrosomal functions may contribute to coordinating chromosome replication and centrosomal function.
Experimental procedures
DNA construction and transfection
FLAG-tagged full-length wild-type or the L313A/I316A (LI/AA) mutant form of the cdc6 open reading frame was cloned into the pTRE2hyg vector (Clontech). Deleted and/or mutated cdc6 DNA were cloned into the p3XFLAG-CMV7 vector (Sigma). GFP-tagged Cdc6(75–366)-PACT, in wild-type or mutant forms, were cloned into the pcDNA5/FRT/TO vector (62). DNA constructs were transfected into cells using Lipofectamine 2000 transfection reagent (Invitrogen).
Short interfering RNA (siRNA) transfection
siRNA oligonucleotides were purchased from ST Pharm. GL3 siRNA, the control, had the following sequence: 5′-CUU ACG CUG AGU ACU UCG ATT-3′. Cdc6 siRNA had the following sequence: 5′-UAA GCC GGA UUC UGC AAG A-3′. siRNA oligonucleotides were transfected into cells using Oligofectamine (Invitrogen).
Cell culture and cell line construction
U2OS and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics (penicillin/streptomycin). To generate the siRNA-resistant Cdc6, site-directed mutagenesis was performed by using the primers 5′-CTG CCT GCT TAA GTC GGA TCC TGC AGG ACC TCA AGA AGG-3′ and 5′-CCT TCT TGA GGT CCT GCA GGA TCC GAC TTA AGC AGG CAG-3′. The Cdc6 siRNA-resistant, FLAG-tagged, wild-type or mutant Cdc6 DNA was cloned into the pTRE2hyg vector (Clontech) and transfected into U2OS Tet-On cells (Clontech). GFP-tagged Cdc6(75–366)-PACT wild-type or mutant forms were transfected into FRT/TO HeLa cells (gift from Dr. H. Lee). Hygromycin-resistant cells were selected by culturing in 200 μg/ml of hygromycin for 2 weeks, and were then used in experiments. Expression of Cdc6 was induced by addition of 2 μg/ml of doxycycline to the culture medium and allowed to incubate for 48 h.
Antibodies
Anti-Cdc6 (sc-13136, Santa Cruz), anti-α-tubulin (ab18251, Sigma), anti-EB1 (AB6057, Millipore), anti-β-tubulin (sc-9104, Santa Cruz), anti-γ-tubulin (T6557, Sigma), anti-FLAG (F1804, Sigma), anti-cyclin E (sc-25303, Santa Cruz), anti-cyclin A (sc-751, Santa Cruz), anti-mCherry (NBP2–25157, Novus Biologicals), and anti-GFP (sc-9996, Santa Cruz) were used as the antibodies. Cep215/CDK5RAP2 (63), Pericentrin (64), and Cep192 (65) antibodies have been described previously.
BrdU incorporation and fluorescence-activated cell sorting (FACS) analysis
BrdU incorporation was performed by pulsing cells with 10 μm BrdU for 45 min and then staining with BrdU Flow Kits (BD Biosciences). Cell cycles were analyzed using the FACS Calibur instrument (BD Biosciences).
Microtubule regrowth assay
The microtubule regrowth assay was performed as previously described (63), with minor modifications. Asynchronously grown cells were incubated for 1 h on ice to depolymerize microtubules. To allow microtubule regrowth, cells were washed with PBS and incubated for 15 s, unless indicated, in fresh medium at 37 °C, followed by fixation in PEM + Fixative buffer (80 mm PIPES, 5 mm EGTA, 1 mm MgCl2, 0.5% Triton X-100, and 4% paraformaldehyde) for 10 min at room temperature. Microtubule regrowth was quantified via immunostaining; the fluorescence intensity of an area of 5 μm diameter around the centrosomes was measured using ImageJ software.
Immunofluorescence microscopy
Asynchronously grown cells, unless indicated otherwise, were grown on coverslips and fixed with cold methanol to detect centrosomal proteins for 10 min. To visualize the microtubule proteins, cells were fixed with PEM + Fixative buffer. The cells were then permeabilized by incubation with 0.5% PBST (PBS containing 0.5% Triton X-100) for 10 min. After a 30-min incubation in blocking solution (PBS containing 3% bovine serum albumin (BSA) and 0.1% Triton X-100), the cells were immunostained with the indicated antibodies. Then, cells were washed three times with 0.1% PBST, incubated with Cy3- or FITC-conjugated anti-rabbit or anti-mouse secondary antibodies, washed three times with 0.1% PBST, and then mounted on glass slides with mounting media (Biomeda Corp.) containing 1 μg/ml of 4′,6-diamidino-2-phenylindole (DAPI, Vectashield).
Unless indicated otherwise, cells were viewed using an Olympus BX51 microscope equipped with 100 × 1.35 NA UPlanSApo oil immersion objective lens (Olympus) and a digital camera (QImaging QICAM Fast 1394). The images were processed in ImagePro 5.0 (Media Cybernetics). ImageJ (National Institutes of Health) was used to measure fluorescence intensities at centrosomes. In each measurement, background signals were subtracted from fluorescence signals from the centrosome.
Live-cell imaging
EB1-GFP-expressing stable U2OS cells grown in 8-well chambered coverglass (Nunc Lab-Tek) were treated with the indicated siRNAs 24 h before being transfected with the indicated DNA constructs. Twenty-four hours after the DNA transfection, the cells on the chambered coverglass were incubated on ice for 1 h to depolymerize microtubules. They were then transferred to the environmental chamber and pre-warmed at 25 °C of Delta Vision. Microtubule regrowth was performed at 25 °C, and the fluorescence of EB1-GFP and mCherry-Cdc6 was observed under a Delta Vision (GE Healthcare Life Sciences), equipped with a camera (CoolSNAP HQ, Roper Scientific) and a 100 × 1.4 NA UPlanSApo oil objective lens (Olympus). Time-lapse images were acquired with an exposure time of 0.4 s at 30-s intervals for ∼4 min. SoftWoRx was used to analyze the obtained images.
The microtubule nucleation rate was determined by counting the number of EB1 comets emerging from the centrosomes at each time point in at least 10 different cells. The rate was presented as the average number of newly nucleated microtubules per minute. The microtubule growth rate was determined by measuring the length of EB1-GFP comets of five microtubules from the centrosomes of at least 10 different cells. The average length of newly nucleated microtubules per minute was presented.
Statistical analysis
Groups were compared using two-tailed Student's t test and Prism software (GraphPad). A p value below 0.05 was considered statistically significant. At least three independent experiments were performed for statistical analyses.
Author contributions
I. L., G. S. K., and J. K. performed the microtubule and centrosome assays and constructed the cell lines. J. S. B. performed cloning and constructed the cell lines. K. R. supervised J. K. D. S. H. prepared the manuscript and is the corresponding author.
Supplementary Material
This work was supported by Basic Research Laboratory Grant NRF-2014R1A4A10052590 through the National Research Foundation of Korea and a BK21 Research Fellowship from the Ministry of Education, Science and Technology, Republic of Korea (to I. L., G. S. K., J. B., and J. K.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S5 and Movies A–D.
- PCM
- pericentriolar material
- CLS
- centrosome localization signal
- LI/AA
- L313A/I316A
- MTOC
- microtubule-organizing center
- pre-RC
- pre-replicative complex
- TuRC
- γ-tubulin ring complexes.
References
- 1. Gould R. R., and Borisy G. G. (1977) The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. J. Cell Biol. 73, 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Paintrand M., Moudjou M., Delacroix H., and Bornens M. (1992) Centrosome organization and centriole architecture: their sensitivity to divalent cations. J. Struct. Biol. 108, 107–128 [DOI] [PubMed] [Google Scholar]
- 3. Bettencourt-Dias M., and Glover D. M. (2007) Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8, 451–463 [DOI] [PubMed] [Google Scholar]
- 4. Lawo S., Hasegan M., Gupta G. D., and Pelletier L. (2012) Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14, 1148–1158 [DOI] [PubMed] [Google Scholar]
- 5. Woodruff J. B., Wueseke O., and Hyman A. A. (2014) Pericentriolar material structure and dynamics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, pii.20130459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vaughan S., and Dawe H. R. (2011) Common themes in centriole and centrosome movements. Trends Cell Biol. 21, 57–66 [DOI] [PubMed] [Google Scholar]
- 7. Zheng Y., Wong M. L., Alberts B., and Mitchison T. (1995) Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature 378, 578–583 [DOI] [PubMed] [Google Scholar]
- 8. Moritz M., Braunfeld M. B., Sedat J. W., Alberts B., and Agard D. A. (1995) Microtubule nucleation by γ-tubulin-containing rings in the centrosome. Nature 378, 638–640 [DOI] [PubMed] [Google Scholar]
- 9. Moritz M., Braunfeld M. B., Guénebaut V., Heuser J., and Agard D. A. (2000) Structure of the γ-tubulin ring complex: a template for microtubule nucleation. Nat. Cell Biol. 2, 365–370 [DOI] [PubMed] [Google Scholar]
- 10. Fontalba A., Paciucci R., Avila J., and Zabala J. C. (1993) Incorporation of tubulin subunits into dimers requires GTP hydrolysis. J. Cell Sci. 106, 627–632 [DOI] [PubMed] [Google Scholar]
- 11. Nogales E., Wolf S. G., and Downing K. H. (1998) Structure of the αβ tubulin dimer by electron crystallography. Nature 391, 199–203 [DOI] [PubMed] [Google Scholar]
- 12. Desai A., and Mitchison T. J. (1997) Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83–117 [DOI] [PubMed] [Google Scholar]
- 13. Drechsel D. N., Hyman A. A., Cobb M. H., and Kirschner M. W. (1992) Modulation of the dynamic instability of tubulin assembly by the microtubule-associated protein tau. Mol. Biol. Cell 3, 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Janke C., and Bulinski J. C. (2011) Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 12, 773–786 [DOI] [PubMed] [Google Scholar]
- 15. Gönczy P. (2004) Centrosomes: hooked on the nucleus. Curr. Biol. 14, R268–R270 [DOI] [PubMed] [Google Scholar]
- 16. Tanaka T. U., and Desai A. (2008) Kinetochore-microtubule interactions: the means to the end. Curr. Opin. Cell Biol. 20, 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Franker M. A., and Hoogenraad C. C. (2013) Microtubule-based transport: basic mechanisms, traffic rules and role in neurological pathogenesis. J. Cell Sci. 126, 2319–2329 [DOI] [PubMed] [Google Scholar]
- 18. Ganguly A., Yang H., Sharma R., Patel K. D., and Cabral F. (2012) The role of microtubules and their dynamics in cell migration. J. Biol. Chem. 287, 43359–43369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siegrist S. E., and Doe C. Q. (2007) Microtubule-induced cortical cell polarity. Genes Dev. 21, 483–496 [DOI] [PubMed] [Google Scholar]
- 20. Fragkos M., Ganier O., Coulombe P., and Méchali M. (2015) DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 16, 360–374 [DOI] [PubMed] [Google Scholar]
- 21. Hook S. S., Lin J. J., and Dutta A. (2007) Mechanisms to control rereplication and implications for cancer. Curr. Opin. Cell Biol. 19, 663–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Petersen B. O., Lukas J., Sørensen C. S., Bartek J., and Helin K. (1999) Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 18, 396–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang W., Wells N. J., and Hunter T. (1999) Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl. Acad. Sci. U.S.A. 96, 6193–6198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paolinelli R., Mendoza-Maldonado R., Cereseto A., and Giacca M. (2009) Acetylation by GCN5 regulates CDC6 phosphorylation in the S phase of the cell cycle. Nat. Struct. Mol. Biol. 16, 412–420 [DOI] [PubMed] [Google Scholar]
- 25. Donovan S., Harwood J., Drury L. S., and Diffley J. F. (1997) Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 94, 5611–5616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Herbig U., Marlar C. A., and Fanning E. (1999) The Cdc6 nucleotide-binding site regulates its activity in DNA replication in human cells. Mol. Biol. Cell 10, 2631–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Randell J. C., Bowers J. L., Rodríguez H. K., and Bell S. P. (2006) Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Mol. Cell 21, 29–39 [DOI] [PubMed] [Google Scholar]
- 28. Weinreich M., Liang C., and Stillman B. (1999) The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin. Proc. Natl. Acad. Sci. U.S.A. 96, 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kan Q., Jinno S., Yamamoto H., Kobayashi K., and Okayama H. (2008) ATP-dependent activation of p21WAF1/CIP1-associated Cdk2 by Cdc6. Proc. Natl. Acad. Sci. U.S.A. 105, 4757–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Uranbileg B., Yamamoto H., Park J. H., Mohanty A. R., Arakawa-Takeuchi S., Jinno S., and Okayama H. (2012) Cdc6 protein activates p27KIP1-bound Cdk2 protein only after the bound p27 protein undergoes C-terminal phosphorylation. J. Biol. Chem. 287, 6275–6283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Niimi S., Arakawa-Takeuchi S., Uranbileg B., Park J. H., Jinno S., and Okayama H. (2012) Cdc6 protein obstructs apoptosome assembly and consequent cell death by forming stable complexes with activated Apaf-1 molecules. J. Biol. Chem. 287, 18573–18583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saha P., Chen J., Thome K. C., Lawlis S. J., Hou Z. H., Hendricks M., Parvin J. D., and Dutta A. (1998) Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol. 18, 2758–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Petersen B. O., Wagener C., Marinoni F., Kramer E. R., Melixetian M., Lazzerini Denchi E., Gieffers C., Matteucci C., Peters J. M., and Helin K. (2000) Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 14, 2330–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clijsters L., and Wolthuis R. (2014) PIP-box-mediated degradation prohibits re-accumulation of Cdc6 during S phase. J. Cell Sci. 127, 1336–1345 [DOI] [PubMed] [Google Scholar]
- 35. Coverley D., Pelizon C., Trewick S., and Laskey R. A. (2000) Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J. Cell Sci. 113, 1929–1938 [DOI] [PubMed] [Google Scholar]
- 36. Kim G. S., Kang J., Bang S. W., and Hwang D. S. (2015) Cdc6 localizes to S- and G2-phase centrosomes in a cell cycle-dependent manner. Biochem. Biophys. Res. Commun. 456, 763–767 [DOI] [PubMed] [Google Scholar]
- 37. Narasimhachar Y., Webster D. R., Gard D. L., and Coué M. (2012) Cdc6 is required for meiotic spindle assembly in Xenopus oocytes. Cell Cycle 11, 524–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shin W., Yu N. K., Kaang B. K., and Rhee K. (2015) The microtubule nucleation activity of centrobin in both the centrosome and cytoplasm. Cell Cycle 14, 1925–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morrison E. E., Wardleworth B. N., Askham J. M., Markham A. F., and Meredith D. M. (1998) EB1, a protein which interacts with the APC tumour suppressor, is associated with the microtubule cytoskeleton throughout the cell cycle. Oncogene 17, 3471–3477 [DOI] [PubMed] [Google Scholar]
- 40. Tirnauer J. S., Grego S., Salmon E. D., and Mitchison T. J. (2002) EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol. Biol. Cell 13, 3614–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gillingham A. K., and Munro S. (2000) The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1, 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matsumoto Y., and Maller J. L. (2004) A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science 306, 885–888 [DOI] [PubMed] [Google Scholar]
- 43. Koff A., Giordano A., Desai D., Yamashita K., Harper J. W., Elledge S., Nishimoto T., Morgan D. O., Franza B. R., and Roberts J. M. (1992) Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science 257, 1689–1694 [DOI] [PubMed] [Google Scholar]
- 44. Pagano M., Pepperkok R., Verde F., Ansorge W., and Draetta G. (1992) Cyclin A is required at two points in the human cell cycle. EMBO J. 11, 961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Delaval B., and Doxsey S. J. (2010) Pericentrin in cellular function and disease. J. Cell Biol. 188, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee K., and Rhee K. (2011) PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J. Cell Biol. 195, 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Doxsey S. J., Stein P., Evans L., Calarco P. D., and Kirschner M. (1994) Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell 76, 639–650 [DOI] [PubMed] [Google Scholar]
- 48. Dictenberg J. B., Zimmerman W., Sparks C. A., Young A., Vidair C., Zheng Y., Carrington W., Fay F. S., and Doxsey S. J. (1998) Pericentrin and γ-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 141, 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gomez-Ferreria M. A., Rath U., Buster D. W., Chanda S. K., Caldwell J. S., Rines D. R., and Sharp D. J. (2007) Human Cep192 is required for mitotic centrosome and spindle assembly. Curr. Biol. 17, 1960–1966 [DOI] [PubMed] [Google Scholar]
- 50. Starita L. M., Machida Y., Sankaran S., Elias J. E., Griffin K., Schlegel B. P., Gygi S. P., and Parvin J. D. (2004) BRCA1-dependent ubiquitination of γ-tubulin regulates centrosome number. Mol. Cell. Biol. 24, 8457–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Park J., and Rhee K. (2013) NEK2 phosphorylation antagonizes the microtubule stabilizing activity of centrobin. Biochem. Biophys. Res. Commun. 431, 302–308 [DOI] [PubMed] [Google Scholar]
- 52. Tsou M. F., and Stearns T. (2006) Mechanism limiting centrosome duplication to once per cell cycle. Nature 442, 947–951 [DOI] [PubMed] [Google Scholar]
- 53. Vitre B. D., and Cleveland D. W. (2012) Centrosomes, chromosome instability (CIN) and aneuploidy. Curr. Opin. Cell Biol. 24, 809–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lacey K. R., Jackson P. K., and Stearns T. (1999) Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. U.S.A. 96, 2817–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nam H. J., and van Deursen J. M. (2014) Cyclin B2 and p53 control proper timing of centrosome separation. Nat. Cell Biol. 16, 538–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hemerly A. S., Prasanth S. G., Siddiqui K., and Stillman B. (2009) Orc1 controls centriole and centrosome copy number in human cells. Science 323, 789–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Prasanth S. G., Prasanth K. V., Siddiqui K., Spector D. L., and Stillman B. (2004) Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 23, 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stuermer A., Hoehn K., Faul T., Auth T., Brand N., Kneissl M., Pütter V., and Grummt F. (2007) Mouse pre-replicative complex proteins colocalise and interact with the centrosome. Eur. J. Cell Biol. 86, 37–50 [DOI] [PubMed] [Google Scholar]
- 59. Ferguson R. L., and Maller J. L. (2008) Cyclin E-dependent localization of MCM5 regulates centrosome duplication. J. Cell Sci. 121, 3224–3232 [DOI] [PubMed] [Google Scholar]
- 60. Ferguson R. L., Pascreau G., and Maller J. L. (2010) The cyclin A centrosomal localization sequence recruits MCM5 and Orc1 to regulate centrosome reduplication. J. Cell Sci. 123, 2743–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lu F., Lan R., Zhang H., Jiang Q., and Zhang C. (2009) Geminin is partially localized to the centrosome and plays a role in proper centrosome duplication. Biol. Cell 101, 273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Han J. S., Vitre B., Fachinetti D., and Cleveland D. W. (2014) Bimodal activation of BubR1 by Bub3 sustains mitotic checkpoint signaling. Proc. Natl. Acad. Sci. U.S.A. 111, E4185–E4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee S., and Rhee K. (2010) CEP215 is involved in the dynein-dependent accumulation of pericentriolar matrix proteins for spindle pole formation. Cell Cycle 9, 774–783 [PubMed] [Google Scholar]
- 64. Kim K., and Rhee K. (2011) The pericentriolar satellite protein CEP90 is crucial for integrity of the mitotic spindle pole. J. Cell Sci. 124, 338–347 [DOI] [PubMed] [Google Scholar]
- 65. Kim S., and Rhee K. (2014) Importance of the CEP215-pericentrin interaction for centrosome maturation during mitosis. PloS One 9, e87016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Borlado L. R., and Méndez J. (2008) CDC6: from DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis 29, 237–243 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.